Intracellular connections between basal bodies promote the coordinated behavior of motile cilia
- PMID: 35767367
- PMCID: PMC9582806
- DOI: 10.1091/mbc.E22-05-0150
Intracellular connections between basal bodies promote the coordinated behavior of motile cilia
Abstract
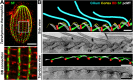
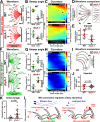
Hydrodynamic flow produced by multiciliated cells is critical for fluid circulation and cell motility. Hundreds of cilia beat with metachronal synchrony for fluid flow. Cilia-driven fluid flow produces extracellular hydrodynamic forces that cause neighboring cilia to beat in a synchronized manner. However, hydrodynamic coupling between neighboring cilia is not the sole mechanism that drives cilia synchrony. Cilia are nucleated by basal bodies (BBs) that link to each other and to the cell's cortex via BB-associated appendages. The intracellular BB and cortical network is hypothesized to synchronize ciliary beating by transmitting cilia coordination cues. The extent of intracellular ciliary connections and the nature of these stimuli remain unclear. Moreover, how BB connections influence the dynamics of individual cilia has not been established. We show by focused ion beam scanning electron microscopy imaging that cilia are coupled both longitudinally and laterally in the ciliate Tetrahymena thermophila by the underlying BB and cortical cytoskeletal network. To visualize the behavior of individual cilia in live, immobilized Tetrahymena cells, we developed
Figures



References
-
- Allen RD (1967). Fine structure, reconstruction and possible functions of components of the cortex of Tetrahymena pyriformis. J Protozool 14, 553–565. - PubMed
-
- Aufderheide KJ (1986). Identification of the basal bodies and kinetodesmal fibers in living cells of Paramecium tetraurelia Sonneborn, 1975 and Paramecium sonneborni Aufderheide, Daggett & Nerad, 1983. J Protozool 33, 77–80. - PubMed
-
- Baena V, Conrad R, Friday P, Fitzgerald E, Kim T, Bernbaum J, Berensmann H, Harned A, Nagashima K, Narayan K (2021). FIB-SEM as a volume electron microscopy approach to study cellular architectures in SARS-CoV-2 and other viral infections: a practical primer for a virologist. Viruses 13, 611. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

