The origin of RNA interference: Adaptive or neutral evolution?
- PMID: 35767561
- PMCID: PMC9275709
- DOI: 10.1371/journal.pbio.3001715
The origin of RNA interference: Adaptive or neutral evolution?
Abstract
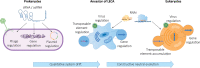
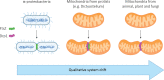
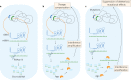
The origin of RNA interference (RNAi) is usually explained by a defense-based hypothesis, in which RNAi evolved as a defense against transposable elements (TEs) and RNA viruses and was already present in the last eukaryotic common ancestor (LECA). However, since RNA antisense regulation and double-stranded RNAs (dsRNAs) are ancient and widespread phenomena, the origin of defensive RNAi should have occurred in parallel with its regulative functions to avoid imbalances in gene regulation. Thus, we propose a neutral evolutionary hypothesis for the origin of RNAi in which qualitative system drift from a prokaryotic antisense RNA gene regulation mechanism leads to the formation of RNAi through constructive neutral evolution (CNE). We argue that RNAi was already present in the ancestor of LECA before the need for a new defense system arose and that its presence helped to shape eukaryotic genomic architecture and stability.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- Gould SJ, Lewontin LC. The spandrels of San Marco and the Panglossian paradigm: a critique of the adaptationist programme. Proc R Soc Lond B Biol Sci [Internet]. 1979. Sep 21;205(1161):581–98. Available from: http://www.royalsocietypublishing.org/doi/10.1098/rspb.1979.0086 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

