Discovering molecular features of intrinsically disordered regions by using evolution for contrastive learning
- PMID: 35767567
- PMCID: PMC9275697
- DOI: 10.1371/journal.pcbi.1010238
Discovering molecular features of intrinsically disordered regions by using evolution for contrastive learning
Abstract
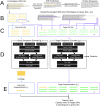
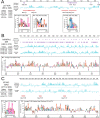
A major challenge to the characterization of intrinsically disordered regions (IDRs), which are widespread in the proteome, but relatively poorly understood, is the identification of molecular features that mediate functions of these regions, such as short motifs, amino acid repeats and physicochemical properties. Here, we introduce a proteome-scale feature discovery approach for IDRs. Our approach, which we call "reverse homology", exploits the principle that important functional features are conserved over evolution. We use this as a contrastive learning signal for deep learning: given a set of homologous IDRs, the neural network has to correctly choose a held-out homolog from another set of IDRs sampled randomly from the proteome. We pair reverse homology with a simple architecture and standard interpretation techniques, and show that the network learns conserved features of IDRs that can be interpreted as motifs, repeats, or bulk features like charge or amino acid propensities. We also show that our model can be used to produce visualizations of what residues and regions are most important to IDR function, generating hypotheses for uncharacterized IDRs. Our results suggest that feature discovery using unsupervised neural networks is a promising avenue to gain systematic insight into poorly understood protein sequences.
Conflict of interest statement
I have read the journal’s policy and the authors of this manuscript have the following competing interests: AMM is a Consultant to Dewpoint Therapeutics Inc.
Figures




References
-
- Lindorff-Larsen K, Kragelund BB. On the potential of machine learning to examine the relationship between sequence, structure, dynamics and function of intrinsically disordered proteins. 2021. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

