Transcriptomic modulation in response to an intoxication with deltamethrin in a population of Triatoma infestans with low resistance to pyrethroids
- PMID: 35767570
- PMCID: PMC9275713
- DOI: 10.1371/journal.pntd.0010060
Transcriptomic modulation in response to an intoxication with deltamethrin in a population of Triatoma infestans with low resistance to pyrethroids
Abstract
Background: Triatoma infestans is the main vector of Chagas disease in the Southern Cone. The resistance to pyrethroid insecticides developed by populations of this species impairs the effectiveness of vector control campaigns in wide regions of Argentina. The study of the global transcriptomic response to pyrethroid insecticides is important to deepen the knowledge about detoxification in triatomines.
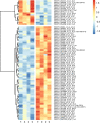
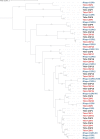
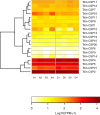
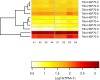
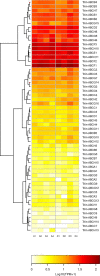
Methodology and findings: We used RNA-Seq to explore the early transcriptomic response after intoxication with deltamethrin in a population of T. infestans which presents low resistance to pyrethroids. We were able to assemble a complete transcriptome of this vector and found evidence of differentially expressed genes belonging to diverse families such as chemosensory and odorant-binding proteins, ABC transporters and heat-shock proteins. Moreover, genes related to transcription and translation, energetic metabolism and cuticle rearrangements were also modulated. Finally, we characterized the repertoire of previously uncharacterized detoxification-related gene families in T. infestans and Rhodnius prolixus.
Conclusions and significance: Our work contributes to the understanding of the detoxification response in vectors of Chagas disease. Given the absence of an annotated genome from T. infestans, the analysis presented here constitutes a resource for molecular and physiological studies in this species. The results increase the knowledge on detoxification processes in vectors of Chagas disease, and provide relevant information to explore undescribed potential insecticide resistance mechanisms in populations of these insects.
Conflict of interest statement
I have read the journal’s policy and the authors of this manuscript have the following competing interests: J.M.L.E, G.F, P.L, G.M.C and S.O are investigators from Consejo Nacional de Ciencia y Tecnologıa (CONICET; https://www.conicet.gov.ar/). L.T. is recipient of a research fellowship from CONICET.
Figures
References
-
- Dias JCP. Southern Cone Initiative for the elimination of domestic populations of Triatoma infestans and the interruption of transfusion Chagas disease: historical aspects, present situation, and perspectives. Mem Inst Oswaldo Cruz. 2007;102: 11–18. doi: 10.1590/s0074-02762007005000092 - DOI - PubMed
-
- Martínez-Barnetche J, Lavore A, Beliera M, Téllez-Sosa J, Zumaya-Estrada FA, Palacio V, et al. Adaptations in energy metabolism and gene family expansions revealed by comparative transcriptomics of three Chagas disease triatomine vectors. BMC Genomics. 2018;19: 296. doi: 10.1186/s12864-018-4696-8 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

