Identification of an integrase-independent pathway of retrotransposition
- PMID: 35767609
- PMCID: PMC11042837
- DOI: 10.1126/sciadv.abm9390
Identification of an integrase-independent pathway of retrotransposition
Abstract
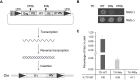
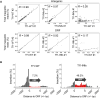
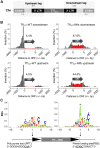
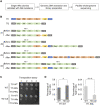
Retroviruses and long terminal repeat retrotransposons rely on integrase (IN) to insert their complementary DNA (cDNA) into the genome of host cells. Nevertheless, in the absence of IN, retroelements can retain notable levels of insertion activity. We have characterized the IN-independent pathway of Tf1 and found that insertion sites had homology to the primers of reverse transcription, which form single-stranded DNAs at the termini of the cDNA. In the absence of IN activity, a similar bias was observed with HIV-1. Our studies showed that the Tf1 insertions result from single-strand annealing, a noncanonical form of homologous recombination mediated by Rad52. By expanding our analysis of insertions to include repeat sequences, we found most formed tandem elements by inserting at preexisting copies of a related transposable element. Unexpectedly, we found that wild-type Tf1 uses the IN-independent pathway as an alternative mode of insertion.
Figures

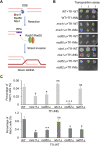
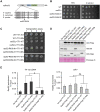
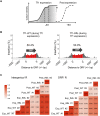

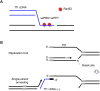
Similar articles
-
Qualitative and Quantitative Assays of Transposition and Homologous Recombination of the Retrotransposon Tf1 in Schizosaccharomyces pombe.Methods Mol Biol. 2016;1400:117-30. doi: 10.1007/978-1-4939-3372-3_8. Methods Mol Biol. 2016. PMID: 26895050 Free PMC article.
-
The removal of RNA primers from DNA synthesized by the reverse transcriptase of the retrotransposon Tf1 is stimulated by Tf1 integrase.J Virol. 2012 Jun;86(11):6222-30. doi: 10.1128/JVI.00009-12. Epub 2012 Apr 4. J Virol. 2012. PMID: 22491446 Free PMC article.
-
cDNA of the yeast retrotransposon Ty5 preferentially recombines with substrates in silent chromatin.Mol Cell Biol. 1999 Jan;19(1):484-94. doi: 10.1128/MCB.19.1.484. Mol Cell Biol. 1999. PMID: 9858572 Free PMC article.
-
"In the beginning": initiation of minus strand DNA synthesis in retroviruses and LTR-containing retrotransposons.Biochemistry. 2003 Dec 16;42(49):14349-55. doi: 10.1021/bi030201q. Biochemistry. 2003. PMID: 14661945 Review.
-
DIRS-1 and the other tyrosine recombinase retrotransposons.Cytogenet Genome Res. 2005;110(1-4):575-88. doi: 10.1159/000084991. Cytogenet Genome Res. 2005. PMID: 16093711 Review.
Cited by
-
Homodecameric Rad52 promotes single-position Rad51 nucleation in homologous recombination.bioRxiv [Preprint]. 2023 Jun 6:2023.02.05.527205. doi: 10.1101/2023.02.05.527205. bioRxiv. 2023. PMID: 36778491 Free PMC article. Preprint.
-
Tandem LTR-retrotransposon structures are common and highly polymorphic in plant genomes.Mob DNA. 2025 Mar 12;16(1):10. doi: 10.1186/s13100-025-00347-y. Mob DNA. 2025. PMID: 40075446 Free PMC article.
-
Association analysis of response to take-all disease with agronomic traits and molecular markers and selection ideal genotypes in bread wheat (Triticum aestivum L.) genotypes.Mol Breed. 2025 Mar 26;45(4):36. doi: 10.1007/s11032-025-01554-4. eCollection 2025 Apr. Mol Breed. 2025. PMID: 40151761
-
Endogenous Caulimovirids: Fossils, Zombies, and Living in Plant Genomes.Biomolecules. 2023 Jul 3;13(7):1069. doi: 10.3390/biom13071069. Biomolecules. 2023. PMID: 37509105 Free PMC article. Review.
-
Transposon Removal Reveals Their Adaptive Fitness Contribution.Genome Biol Evol. 2024 Feb 1;16(2):evae010. doi: 10.1093/gbe/evae010. Genome Biol Evol. 2024. PMID: 38245838 Free PMC article.
References
-
- Deeks S. G., Overbaugh J., Phillips A., Buchbinder S., HIV infection. Nat. Rev. Dis. Primers. 1, 15035 (2015). - PubMed
-
- Saag M. S., Gandhi R. T., Hoy J. F., Landovitz R. J., Thompson M. A., Sax P. E., Smith D. M., Benson C. A., Buchbinder S. P., del Rio C., Eron J. J. Jr., Fätkenheuer G., Günthard H. F., Molina J. M., Jacobsen D. M., Volberding P. A., Antiretroviral drugs for treatment and prevention of hiv infection in Adults: 2020 recommendations of the international antiviral society-USA panel. JAMA 324, 1651–1669 (2020). - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

