In vivo multidimensional CRISPR screens identify Lgals2 as an immunotherapy target in triple-negative breast cancer
- PMID: 35767614
- PMCID: PMC9242595
- DOI: 10.1126/sciadv.abl8247
In vivo multidimensional CRISPR screens identify Lgals2 as an immunotherapy target in triple-negative breast cancer
Abstract
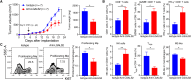
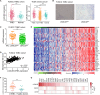
Immune checkpoint inhibitors exhibit limited response rates in patients with triple-negative breast cancer (TNBC), suggesting that additional immune escape mechanisms may exist. Here, we performed two-step customized in vivo CRISPR screens targeting disease-related immune genes using different mouse models with multidimensional immune-deficiency characteristics. In vivo screens characterized gene functions in the different tumor microenvironments and recovered canonical immunotherapy targets such as Ido1. In addition, functional screening and transcriptomic analysis identified Lgals2 as a candidate regulator in TNBC involving immune escape. Mechanistic studies demonstrated that tumor cell-intrinsic Lgals2 induced the increased number of tumor-associated macrophages, as well as the M2-like polarization and proliferation of macrophages through the CSF1/CSF1R axis, which resulted in the immunosuppressive nature of the TNBC microenvironment. Blockade of LGALS2 using an inhibitory antibody successfully arrested tumor growth and reversed the immune suppression. Collectively, our results provide a theoretical basis for LGALS2 as a potential immunotherapy target in TNBC.
Figures






References
-
- Bray F., Ferlay J., Soerjomataram I., Siegel R. L., Torre L. A., Jemal A., Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 68, 394–424 (2018). - PubMed
-
- Denkert C., Liedtke C., Tutt A., von Minckwitz G., Molecular alterations in triple-negative breast cancer—The road to new treatment strategies. Lancet 389, 2430–2442 (2017). - PubMed
-
- Wilky B. A., Immune checkpoint inhibitors: The linchpins of modern immunotherapy. Immunol. Rev. 290, 6–23 (2019). - PubMed
-
- Schmid P., Rugo H. S., Adams S., Schneeweiss A., Barrios C. H., Iwata H., Dieras V., Henschel V., Molinero L., Chui S. Y., Maiya V., Husain A., Winer E. P., Loi S., Emens L. A.; IMpassion130 Investigators , Atezolizumab plus nab-paclitaxel as first-line treatment for unresectable, locally advanced or metastatic triple-negative breast cancer (IMpassion130): Updated efficacy results from a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol. 21, 44–59 (2020). - PubMed
-
- Schmid P., Cortes J., Pusztai L., McArthur H., Kummel S., Bergh J., Denkert C., Park Y. H., Hui R., Harbeck N., Takahashi M., Foukakis T., Fasching P. A., Cardoso F., Untch M., Jia L., Karantza V., Zhao J., Aktan G., Dent R., O’Shaughnessy J.; KEYNOTE-522 Investigators , Pembrolizumab for early triple-negative breast cancer. N. Engl. J. Med. 382, 810–821 (2020). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

