Fragment Screening Yields a Small-Molecule Stabilizer of 14-3-3 Dimers That Modulates Client Protein Interactions
- PMID: 35767695
- PMCID: PMC9543038
- DOI: 10.1002/cbic.202200178
Fragment Screening Yields a Small-Molecule Stabilizer of 14-3-3 Dimers That Modulates Client Protein Interactions
Abstract
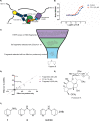
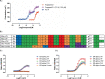
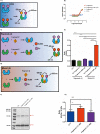
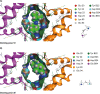
The development of protein-protein interaction (PPI) inhibitors has been a successful strategy in drug development. However, the identification of PPI stabilizers has proven much more challenging. Here we report a fragment-based drug screening approach using the regulatory hub-protein 14-3-3 as a platform for identifying PPI stabilizers. A homogenous time-resolved FRET assay was used to monitor stabilization of 14-3-3/peptide binding using the known interaction partner estrogen receptor alpha. Screening of an in-house fragment library identified fragment 2 (VUF15640) as a putative PPI stabilizer capable of cooperatively stabilizing 14-3-3 PPIs in a cooperative fashion with Fusicoccin-A. Mechanistically, fragment 2 appears to enhance 14-3-3 dimerization leading to increased client-protein binding. Functionally, fragment 2 enhanced potency of 14-3-3 in a cell-free system inhibiting the enzyme activity of the nitrate reductase. In conclusion, we identified a general PPI stabilizer targeting 14-3-3, which could be used as a tool compound for investigating 14-3-3 client protein interactions.
Keywords: drug discovery; estrogen receptor alpha; fragments; protein-protein interactions; stabilizers.
© 2022 The Authors. ChemBioChem published by Wiley-VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

