Pervasive translation of circular RNAs driven by short IRES-like elements
- PMID: 35768398
- PMCID: PMC9242994
- DOI: 10.1038/s41467-022-31327-y
Pervasive translation of circular RNAs driven by short IRES-like elements
Abstract
Some circular RNAs (circRNAs) were found to be translated through IRES-driven mechanism, however the scope and functions of circRNA translation are unclear because endogenous IRESs are rare. To determine the prevalence and mechanism of circRNA translation, we develop a cell-based system to screen random sequences and identify 97 overrepresented hexamers that drive cap-independent circRNA translation. These IRES-like short elements are significantly enriched in endogenous circRNAs and sufficient to drive circRNA translation. We further identify multiple trans-acting factors that bind these IRES-like elements to initiate translation. Using mass-spectrometry data, hundreds of circRNA-coded peptides are identified, most of which have low abundance due to rapid degradation. As judged by mass-spectrometry, 50% of translatable endogenous circRNAs undergo rolling circle translation, several of which are experimentally validated. Consistently, mutations of the IRES-like element in one circRNA reduce its translation. Collectively, our findings suggest a pervasive translation of circRNAs, providing profound implications in translation control.
© 2022. The Author(s).
Conflict of interest statement
Z.W. and Y.Y. has co-founded a company, CirCode Biotech, to commercialize the application of circular RNA as template of protein production/expression, and applied a patent to use circRNA as a gene expression platform. The other authors declare no competing interests.
Figures
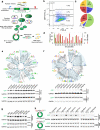
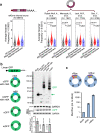
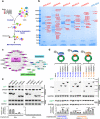

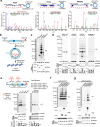
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

