Reply to: Pitfalls in using phenanthroline to study the causal relationship between promoter nucleosome acetylation and transcription
- PMID: 35768425
- PMCID: PMC9243053
- DOI: 10.1038/s41467-022-30351-2
Reply to: Pitfalls in using phenanthroline to study the causal relationship between promoter nucleosome acetylation and transcription
Conflict of interest statement
The authors declare no competing interests.
Figures
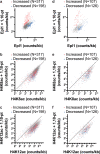
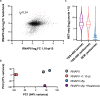
Comment on
-
Transcription shapes genome-wide histone acetylation patterns.Nat Commun. 2021 Jan 11;12(1):210. doi: 10.1038/s41467-020-20543-z. Nat Commun. 2021. PMID: 33431884 Free PMC article.
-
Pitfalls in using phenanthroline to study the causal relationship between promoter nucleosome acetylation and transcription.Nat Commun. 2022 Jun 29;13(1):3726. doi: 10.1038/s41467-022-30350-3. Nat Commun. 2022. PMID: 35768402 Free PMC article. No abstract available.
Similar articles
-
Pitfalls in using phenanthroline to study the causal relationship between promoter nucleosome acetylation and transcription.Nat Commun. 2022 Jun 29;13(1):3726. doi: 10.1038/s41467-022-30350-3. Nat Commun. 2022. PMID: 35768402 Free PMC article. No abstract available.
-
Histone H3 acetylated at lysine 9 in promoter is associated with low nucleosome density in the vicinity of transcription start site in human cell.Chromosome Res. 2006;14(2):203-11. doi: 10.1007/s10577-006-1036-7. Epub 2006 Mar 17. Chromosome Res. 2006. PMID: 16544193
-
Acetylation of nucleosomal histones by p300 facilitates transcription from tax-responsive human T-cell leukemia virus type 1 chromatin template.Mol Cell Biol. 2002 Jul;22(13):4450-62. doi: 10.1128/MCB.22.13.4450-4462.2002. Mol Cell Biol. 2002. PMID: 12052856 Free PMC article.
-
25 years after the nucleosome model: chromatin modifications.Trends Biochem Sci. 2000 Dec;25(12):619-23. doi: 10.1016/s0968-0004(00)01718-7. Trends Biochem Sci. 2000. PMID: 11116189 Review.
-
Prenucleosomes and Active Chromatin.Cold Spring Harb Symp Quant Biol. 2015;80:65-72. doi: 10.1101/sqb.2015.80.027300. Epub 2016 Jan 14. Cold Spring Harb Symp Quant Biol. 2015. PMID: 26767995 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

