WASp modulates RPA function on single-stranded DNA in response to replication stress and DNA damage
- PMID: 35768435
- PMCID: PMC9243104
- DOI: 10.1038/s41467-022-31415-z
WASp modulates RPA function on single-stranded DNA in response to replication stress and DNA damage
Abstract
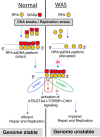
Perturbation in the replication-stress response (RSR) and DNA-damage response (DDR) causes genomic instability. Genomic instability occurs in Wiskott-Aldrich syndrome (WAS), a primary immunodeficiency disorder, yet the mechanism remains largely uncharacterized. Replication protein A (RPA), a single-strand DNA (ssDNA) binding protein, has key roles in the RSR and DDR. Here we show that human WAS-protein (WASp) modulates RPA functions at perturbed replication forks (RFs). Following genotoxic insult, WASp accumulates at RFs, associates with RPA, and promotes RPA:ssDNA complexation. WASp deficiency in human lymphocytes destabilizes RPA:ssDNA-complexes, impairs accumulation of RPA, ATR, ETAA1, and TOPBP1 at genotoxin-perturbed RFs, decreases CHK1 activation, and provokes global RF dysfunction. las17 (yeast WAS-homolog)-deficient S. cerevisiae also show decreased ScRPA accumulation at perturbed RFs, impaired DNA recombination, and increased frequency of DNA double-strand break (DSB)-induced single-strand annealing (SSA). Consequently, WASp (or Las17)-deficient cells show increased frequency of DSBs upon genotoxic insult. Our study reveals an evolutionarily conserved, essential role of WASp in the DNA stress-resolution pathway, such that WASp deficiency provokes RPA dysfunction-coupled genomic instability.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
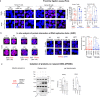

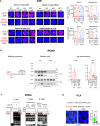

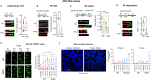
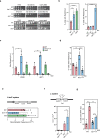
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

