CTCF organizes inter-A compartment interactions through RYBP-dependent phase separation
- PMID: 35768498
- PMCID: PMC9343660
- DOI: 10.1038/s41422-022-00676-0
CTCF organizes inter-A compartment interactions through RYBP-dependent phase separation
Abstract
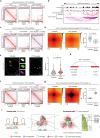
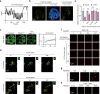
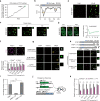
Chromatin is spatially organized into three-dimensional structures at different levels including A/B compartments, topologically associating domains and loops. The canonical CTCF-mediated loop extrusion model can explain the formation of loops. However, the organization mechanisms underlying long-range chromatin interactions such as interactions between A-A compartments are still poorly understood. Here we show that different from the canonical loop extrusion model, RYBP-mediated phase separation of CTCF organizes inter-A compartment interactions. Based on this model, we designed and verified an induced CTCF phase separation system in embryonic stem cells (ESCs), which facilitated inter-A compartment interactions, improved self-renewal of ESCs and inhibited their differentiation toward neural progenitor cells. These findings support a novel and non-canonical role of CTCF in organizing long-range chromatin interactions via phase separation.
© 2022. The Author(s) under exclusive licence to Center for Excellence in Molecular Cell Science, CAS.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

