Enteric viruses replicate in salivary glands and infect through saliva
- PMID: 35768512
- PMCID: PMC9243862
- DOI: 10.1038/s41586-022-04895-8
Enteric viruses replicate in salivary glands and infect through saliva
Abstract
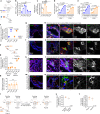
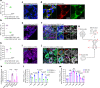
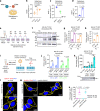
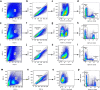
Enteric viruses like norovirus, rotavirus and astrovirus have long been accepted as spreading in the population through fecal-oral transmission: viruses are shed into feces from one host and enter the oral cavity of another, bypassing salivary glands (SGs) and reaching the intestines to replicate, be shed in feces and repeat the transmission cycle1. Yet there are viruses (for example, rabies) that infect the SGs2,3, making the oral cavity one site of replication and saliva one conduit of transmission. Here we report that enteric viruses productively and persistently infect SGs, reaching titres comparable to those in the intestines. We demonstrate that enteric viruses get released into the saliva, identifying a second route of viral transmission. This is particularly significant for infected infants, whose saliva directly transmits enteric viruses to their mothers' mammary glands through backflow during suckling. This sidesteps the conventional gut-mammary axis route4 and leads to a rapid surge in maternal milk secretory IgA antibodies5,6. Lastly, we show that SG-derived spheroids7 and cell lines8 can replicate and propagate enteric viruses, generating a scalable and manageable system of production. Collectively, our research uncovers a new transmission route for enteric viruses with implications for therapeutics, diagnostics and importantly sanitation measures to prevent spread through saliva.
© 2022. This is a U.S. Government work and not under copyright protection in the US; foreign copyright protection may apply.
Conflict of interest statement
The authors declare no competing interests.
Figures



Comment in
-
Norovirus from the mouths of babes.Nature. 2022 Jul;607(7918):247-248. doi: 10.1038/d41586-022-01731-x. Nature. 2022. PMID: 35768603 No abstract available.
References
-
- Posse, J. L., Dios, P. D. & Scully, C. in Saliva Protection and Transmissible Diseases (eds Posse, J. L., Dios, P. D. & Scully, C) 53–92 (Academic Press, 2017).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

