Survival-related indicators ALOX12B and SPRR1A are associated with DNA damage repair and tumor microenvironment status in HPV 16-negative head and neck squamous cell carcinoma patients
- PMID: 35768785
- PMCID: PMC9241267
- DOI: 10.1186/s12885-022-09722-x
Survival-related indicators ALOX12B and SPRR1A are associated with DNA damage repair and tumor microenvironment status in HPV 16-negative head and neck squamous cell carcinoma patients
Erratum in
-
Correction: Survival-related indicators ALOX12B and SPRR1A are associated with DNA damage repair and tumor microenvironment status in HPV 16-negative head and neck squamous cell carcinoma patients.BMC Cancer. 2022 Jul 11;22(1):755. doi: 10.1186/s12885-022-09865-x. BMC Cancer. 2022. PMID: 35820802 Free PMC article. No abstract available.
Abstract
Objectives: To investigate prognostic-related gene signature based on DNA damage repair and tumor microenvironment statue in human papillomavirus 16 negative (HPV16-) head and neck squamous cell carcinoma (HNSCC).
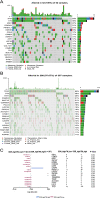
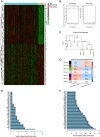
Methods: For the RNA-sequence matrix in HPV16- HNSCC in the Cancer Genome Atlas (TCGA) cohort, the DNA damage response (DDR) and tumor microenvironment (TM) status of each patient sample was estimated by using the ssGSEA algorithm. Through bioinformatics analysis in DDR_high/TM_high (n = 311) and DDR_high/TM_low (n = 53) groups, a survival-related gene signature was selected in the TCGA cohort. Two independent external validation cohorts (GSE65858 (n = 210) and GSE41613 (n = 97)) with HPV16- HNSCC patients validated the gene signature. Correlations among the clinical-related hub differentially expressed genes (DEGs) and infiltrated immunocytes were explored with the TIMER2.0 server. Drug screening based on hub DEGs was performed using the CellMiner and GSCALite databases. The loss-of-function studies were used to evaluate the effect of screened survival-related gene on the motility of HPV- HNSCC cells in vitro.
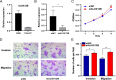
Results: A high DDR level (P = 0.025) and low TM score (P = 0.012) were independent risk factors for HPV16- HNSCC. Downregulated expression of ALOX12B or SPRR1A was associated with poor survival rate and advanced cancer stages. The pathway enrichment analysis showed the DDR_high/TM_low samples were enriched in glycosphingolipid biosynthesis-lacto and neolacto series, glutathione metabolism, platinum drug resistance, and ferroptosis pathways, while the DDR_high/TM_low samples were enriched in Th17 cell differentiation, Neutrophil extracellular trap formation, PD - L1 expression and PD - 1 checkpoint pathway in cancer. Notably, the expression of ALOX12B and SPRR1A were negatively correlated with cancer-associated fibroblasts (CAFs) infiltration and CAFs downstream effectors. Sensitivity to specific chemotherapy regimens can be derived from gene expressions. In addition, ALOX12B and SPRR1A expression was associated with the mRNA expression of insulin like growth factor 1 receptor (IGF1R), AKT serine/threonine kinase 1 (AKT1), mammalian target of rapamycin (MTOR), and eukaryotic translation initiation factor 4E binding protein 1 (EIF4EBP1) in HPV negative HNSCC. Down-regulation of ALOX12B promoted HPV- HNSCC cells migration and invasion in vitro.
Conclusions: ALOX12B and SPRR1A served as a gene signature for overall survival in HPV16- HNSCC patients, and correlated with the amount of infiltrated CAFs. The specific drug pattern was determined by the gene signature.
Keywords: Cancer-associated fibroblasts; DNA damage response; Head and neck squamous cell carcinoma; Human papilloma virus 16-negative; Tumor microenvironment.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures







Similar articles
-
Identification and validation of a prognostic signature of autophagy, apoptosis and pyroptosis-related genes for head and neck squamous cell carcinoma: to imply therapeutic choices of HPV negative patients.Front Immunol. 2023 Jan 10;13:1100417. doi: 10.3389/fimmu.2022.1100417. eCollection 2022. Front Immunol. 2023. PMID: 36703967 Free PMC article.
-
EGFR overexpression increases radiotherapy response in HPV-positive head and neck cancer through inhibition of DNA damage repair and HPV E6 downregulation.Cancer Lett. 2021 Feb 1;498:80-97. doi: 10.1016/j.canlet.2020.10.035. Epub 2020 Oct 31. Cancer Lett. 2021. PMID: 33137407
-
The frequency of high-risk human papillomavirus types, HPV16 lineages, and their relationship with p16INK4a and NF-κB expression in head and neck squamous cell carcinomas in Southwestern Iran.Braz J Microbiol. 2021 Mar;52(1):195-206. doi: 10.1007/s42770-020-00391-1. Epub 2020 Nov 9. Braz J Microbiol. 2021. PMID: 33169334 Free PMC article.
-
Programmed Death-1/Programmed Death-Ligand 1-Axis Blockade in Recurrent or Metastatic Head and Neck Squamous Cell Carcinoma Stratified by Human Papillomavirus Status: A Systematic Review and Meta-Analysis.Front Immunol. 2021 Apr 7;12:645170. doi: 10.3389/fimmu.2021.645170. eCollection 2021. Front Immunol. 2021. PMID: 33897693 Free PMC article.
-
Potential Transcript-Based Biomarkers Predicting Clinical Outcomes of HPV-Positive Head and Neck Squamous Cell Carcinoma Patients.Cells. 2024 Jun 26;13(13):1107. doi: 10.3390/cells13131107. Cells. 2024. PMID: 38994960 Free PMC article. Review.
Cited by
-
Analysis of Selected Small Proline-Rich Proteins in Tissue Homogenates from Samples of Head and Neck Squamous Cell Carcinoma.Diagnostics (Basel). 2025 Jun 26;15(13):1633. doi: 10.3390/diagnostics15131633. Diagnostics (Basel). 2025. PMID: 40647632 Free PMC article.
-
Exploring beyond Common Cell Death Pathways in Oral Cancer: A Systematic Review.Biology (Basel). 2024 Feb 6;13(2):103. doi: 10.3390/biology13020103. Biology (Basel). 2024. PMID: 38392321 Free PMC article. Review.
-
Correction: Survival-related indicators ALOX12B and SPRR1A are associated with DNA damage repair and tumor microenvironment status in HPV 16-negative head and neck squamous cell carcinoma patients.BMC Cancer. 2022 Jul 11;22(1):755. doi: 10.1186/s12885-022-09865-x. BMC Cancer. 2022. PMID: 35820802 Free PMC article. No abstract available.
-
Tumor Microenvironment as a Therapeutic Target in Melanoma Treatment.Cancers (Basel). 2023 Jun 11;15(12):3147. doi: 10.3390/cancers15123147. Cancers (Basel). 2023. PMID: 37370757 Free PMC article. Review.
-
Single-cell RNA sequencing reveals potential therapeutic targets in the tumor microenvironment of lung squamous cell carcinoma.Sci Rep. 2025 Mar 26;15(1):10374. doi: 10.1038/s41598-025-93916-3. Sci Rep. 2025. PMID: 40140461 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

