Comprehensive analysis of CXCR family members in lung adenocarcinoma with prognostic values
- PMID: 35768814
- PMCID: PMC9245315
- DOI: 10.1186/s12890-022-02051-6
Comprehensive analysis of CXCR family members in lung adenocarcinoma with prognostic values
Abstract
Background: The expression profiles and molecular mechanisms of CXC chemokine receptors (CXCRs) in Lung adenocarcinoma (LUAD) have been extensively explored. However, the comprehensive prognostic values of CXCR members in LUAD have not yet been clearly identified.
Methods: Multiple available datasets, including Oncomine datasets, the cancer genome atlas (TCGA), HPA platform, GeneMANIA platform, DAVID platform and the tumor immune estimation resource (TIMER) were used to detect the expression of CXCRs in LUAD, as well as elucidate the significance and value of novel CXCRs-associated genes and signaling pathways in LUAD.
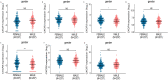
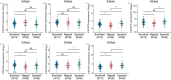
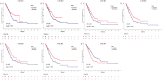
Results: The mRNA and/or protein expression of CXCR1, CXCR2, CXCR3, CXCR4, CXCR5 and CXCR6 displayed predominantly decreased in LUAD tissues as compared to normal tissues. On the contrary, compared with the normal tissues, the expression of CXCR7 was significantly increased in LUAD tissues. Subsequently, we constructed a network including CXCR family members and their 20 related genes, and the related GO functions assay showed that CXCRs connected with these genes participated in the process of LUAD through several signal pathways including Chemokine signaling pathway, Cytokine-cytokine receptor interaction and Neuroactive ligand-receptor interaction. TCGA and Timer platform revealed that the mRNA expression of CXCR family members was significantly related to individual cancer stages, cancer subtypes, patient's gender and the immune infiltration level. Finally, survival analysis showed that low mRNA expression levels of CXCR2 (HR = 0.661, and Log-rank P = 1.90e-02), CXCR3 (HR = 0.674, and Log-rank P = 1.00e-02), CXCR4 (HR = 0.65, and Log-rank P = 5.01e-03), CXCR5 (HR = 0.608, and Log-rank P = 4.80e-03) and CXCR6 (HR = 0.622, and Log-rank P = 1.85e-03) were significantly associated with shorter overall survival (OS), whereas high CXCR7 mRNA expression (HR = 1.604, and Log-rank P = 4.27e-03) was extremely related with shorter OS in patients.
Conclusion: Our findings from public databases provided a unique insight into expression characteristics and prognostic values of CXCR members in LUAD, which would be benefit for the understanding of pathogenesis, diagnosis, prognosis prediction and targeted treatment in LUAD.
Keywords: CXC chemokine receptors (CXCRs); Lung adenocarcinoma (LUAD); Prognostic values.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no conflicts of interest.
Figures





References
-
- Travis WD, Brambilla E, Noguchi M, et al. International association for the study of lung cancer/American thoracic society/European respiratory society: international multidisciplinary classification of lung adenocarcinoma: executive summary. Proc Am Thorac Soc. 2011;8(5):381–385. doi: 10.1513/pats.201107-042ST. - DOI - PubMed
MeSH terms
Substances
Grants and funding
- grant no. 81272854/The present study was supported by the National Natural Science Foundation of China
- grant no. 81272854/The present study was supported by the National Natural Science Foundation of China
- grant no. 81272854/The present study was supported by the National Natural Science Foundation of China
- grant no. 81272854/The present study was supported by the National Natural Science Foundation of China
- grant no. 81272854/The present study was supported by the National Natural Science Foundation of China
- grant no. 81272854/The present study was supported by the National Natural Science Foundation of China
- grant no. 81272854/The present study was supported by the National Natural Science Foundation of China
- grant no. 81272854/The present study was supported by the National Natural Science Foundation of China
- grant no. 81272854/The present study was supported by the National Natural Science Foundation of China
- grant no. 81272854/The present study was supported by the National Natural Science Foundation of China
- grant no. 81272854/The present study was supported by the National Natural Science Foundation of China
- grant no. 81272854/The present study was supported by the National Natural Science Foundation of China
- grant no. 81272854/The present study was supported by the National Natural Science Foundation of China
- grant no. 81272854/The present study was supported by the National Natural Science Foundation of China
- grant no. ZD2019H008/Key Projects of Natural Science Foundation of Heilongjiang Province
- grant no. ZD2019H008/Key Projects of Natural Science Foundation of Heilongjiang Province
- grant no. ZD2019H008/Key Projects of Natural Science Foundation of Heilongjiang Province
- grant no. ZD2019H008/Key Projects of Natural Science Foundation of Heilongjiang Province
- grant no. ZD2019H008/Key Projects of Natural Science Foundation of Heilongjiang Province
- grant no. ZD2019H008/Key Projects of Natural Science Foundation of Heilongjiang Province
- grant no. ZD2019H008/Key Projects of Natural Science Foundation of Heilongjiang Province
- grant no. ZD2019H008/Key Projects of Natural Science Foundation of Heilongjiang Province
- grant no. ZD2019H008/Key Projects of Natural Science Foundation of Heilongjiang Province
- grant no. ZD2019H008/Key Projects of Natural Science Foundation of Heilongjiang Province
- grant no. ZD2019H008/Key Projects of Natural Science Foundation of Heilongjiang Province
- grant no. ZD2019H008/Key Projects of Natural Science Foundation of Heilongjiang Province
- grant no. ZD2019H008/Key Projects of Natural Science Foundation of Heilongjiang Province
- grant no. ZD2019H008/Key Projects of Natural Science Foundation of Heilongjiang Province
- grant no. 2019-KYYWF-1334/Excellent Innovation Team Construction Project of Basic Scientific Research Business Fee of Provincial Colleges and Universities in Heilongjiang Province
- grant no. 2019-KYYWF-1334/Excellent Innovation Team Construction Project of Basic Scientific Research Business Fee of Provincial Colleges and Universities in Heilongjiang Province
- grant no. 2019-KYYWF-1334/Excellent Innovation Team Construction Project of Basic Scientific Research Business Fee of Provincial Colleges and Universities in Heilongjiang Province
- grant no. 2019-KYYWF-1334/Excellent Innovation Team Construction Project of Basic Scientific Research Business Fee of Provincial Colleges and Universities in Heilongjiang Province
- grant no. 2019-KYYWF-1334/Excellent Innovation Team Construction Project of Basic Scientific Research Business Fee of Provincial Colleges and Universities in Heilongjiang Province
- grant no. 2019-KYYWF-1334/Excellent Innovation Team Construction Project of Basic Scientific Research Business Fee of Provincial Colleges and Universities in Heilongjiang Province
- grant no. 2019-KYYWF-1334/Excellent Innovation Team Construction Project of Basic Scientific Research Business Fee of Provincial Colleges and Universities in Heilongjiang Province
- grant no. 2019-KYYWF-1334/Excellent Innovation Team Construction Project of Basic Scientific Research Business Fee of Provincial Colleges and Universities in Heilongjiang Province
- grant no. 2019-KYYWF-1334/Excellent Innovation Team Construction Project of Basic Scientific Research Business Fee of Provincial Colleges and Universities in Heilongjiang Province
- grant no. 2019-KYYWF-1334/Excellent Innovation Team Construction Project of Basic Scientific Research Business Fee of Provincial Colleges and Universities in Heilongjiang Province
- grant no. 2019-KYYWF-1334/Excellent Innovation Team Construction Project of Basic Scientific Research Business Fee of Provincial Colleges and Universities in Heilongjiang Province
- grant no. 2019-KYYWF-1334/Excellent Innovation Team Construction Project of Basic Scientific Research Business Fee of Provincial Colleges and Universities in Heilongjiang Province
- grant no. 2019-KYYWF-1334/Excellent Innovation Team Construction Project of Basic Scientific Research Business Fee of Provincial Colleges and Universities in Heilongjiang Province
- grant no. 2019-KYYWF-1334/Excellent Innovation Team Construction Project of Basic Scientific Research Business Fee of Provincial Colleges and Universities in Heilongjiang Province
- grant no. UNPYSCT-2020054/Young innovative talents training project of regular undergraduate colleges and universities in Heilongjiang Province
- grant no. UNPYSCT-2020054/Young innovative talents training project of regular undergraduate colleges and universities in Heilongjiang Province
- grant no. UNPYSCT-2020054/Young innovative talents training project of regular undergraduate colleges and universities in Heilongjiang Province
- grant no. UNPYSCT-2020054/Young innovative talents training project of regular undergraduate colleges and universities in Heilongjiang Province
- grant no. UNPYSCT-2020054/Young innovative talents training project of regular undergraduate colleges and universities in Heilongjiang Province
- grant no. UNPYSCT-2020054/Young innovative talents training project of regular undergraduate colleges and universities in Heilongjiang Province
- grant no. UNPYSCT-2020054/Young innovative talents training project of regular undergraduate colleges and universities in Heilongjiang Province
- grant no. UNPYSCT-2020054/Young innovative talents training project of regular undergraduate colleges and universities in Heilongjiang Province
- grant no. UNPYSCT-2020054/Young innovative talents training project of regular undergraduate colleges and universities in Heilongjiang Province
- grant no. UNPYSCT-2020054/Young innovative talents training project of regular undergraduate colleges and universities in Heilongjiang Province
- grant no. UNPYSCT-2020054/Young innovative talents training project of regular undergraduate colleges and universities in Heilongjiang Province
- grant no. UNPYSCT-2020054/Young innovative talents training project of regular undergraduate colleges and universities in Heilongjiang Province
- grant no. UNPYSCT-2020054/Young innovative talents training project of regular undergraduate colleges and universities in Heilongjiang Province
- grant no. UNPYSCT-2020054/Young innovative talents training project of regular undergraduate colleges and universities in Heilongjiang Province
- JMSUBZ2019-01/Jiamusi University doctoral Special Research Fund launch project
- JMSUBZ2019-01/Jiamusi University doctoral Special Research Fund launch project
- JMSUBZ2019-01/Jiamusi University doctoral Special Research Fund launch project
- JMSUBZ2019-01/Jiamusi University doctoral Special Research Fund launch project
- JMSUBZ2019-01/Jiamusi University doctoral Special Research Fund launch project
- JMSUBZ2019-01/Jiamusi University doctoral Special Research Fund launch project
- JMSUBZ2019-01/Jiamusi University doctoral Special Research Fund launch project
- JMSUBZ2019-01/Jiamusi University doctoral Special Research Fund launch project
- JMSUBZ2019-01/Jiamusi University doctoral Special Research Fund launch project
- JMSUBZ2019-01/Jiamusi University doctoral Special Research Fund launch project
- JMSUBZ2019-01/Jiamusi University doctoral Special Research Fund launch project
- JMSUBZ2019-01/Jiamusi University doctoral Special Research Fund launch project
- JMSUBZ2019-01/Jiamusi University doctoral Special Research Fund launch project
- JMSUBZ2019-01/Jiamusi University doctoral Special Research Fund launch project
- No.19-112-4-092/Shenyang Science and Technology Plan Project
- No.19-112-4-092/Shenyang Science and Technology Plan Project
- No.19-112-4-092/Shenyang Science and Technology Plan Project
- No.19-112-4-092/Shenyang Science and Technology Plan Project
- No.19-112-4-092/Shenyang Science and Technology Plan Project
- No.19-112-4-092/Shenyang Science and Technology Plan Project
- No.19-112-4-092/Shenyang Science and Technology Plan Project
- No.19-112-4-092/Shenyang Science and Technology Plan Project
- No.19-112-4-092/Shenyang Science and Technology Plan Project
- No.19-112-4-092/Shenyang Science and Technology Plan Project
- No.19-112-4-092/Shenyang Science and Technology Plan Project
- No.19-112-4-092/Shenyang Science and Technology Plan Project
- No.19-112-4-092/Shenyang Science and Technology Plan Project
- No.19-112-4-092/Shenyang Science and Technology Plan Project
LinkOut - more resources
Full Text Sources
Medical

