Composition and Ecological Roles of the Core Microbiome along the Abyssal-Hadal Transition Zone Sediments of the Mariana Trench
- PMID: 35768947
- PMCID: PMC9241748
- DOI: 10.1128/spectrum.01988-21
Composition and Ecological Roles of the Core Microbiome along the Abyssal-Hadal Transition Zone Sediments of the Mariana Trench
Abstract
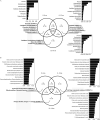
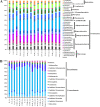
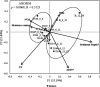
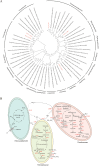
The unique geological features of hadal trenches are known to influence both the structure and ecological function of microbial communities. It is also well known that heterotrophs and chemoautotrophs dominate the hadal and abyssal pelagic zones, respectively. Here, a metagenomic investigation was conducted on sediment samples obtained from the abyssal-hadal transition zone in the Mariana Trench to gain a better understanding of the general diversity and potential function of the core microbiome in this zone. A high level of cosmopolitanism existed in the core microbiome referred from a high community similarity among different stations. Niche differentiation along the fine-scale of different sediment layers was observed, especially for major archaeal groups, largely due to sediment depth and the source of organic matter. A prevalence of nitrogen biogeochemical cycles driven by various nitrifying groups with the capability of dark carbon fixation in the abyssal-hadal biosphere was also demonstrated. The predominance of heterotrophic over chemolithoautotrophic pathways in this transition zone was found, and a high abundance of genes related to respiration and carbon fixation (i.e., the intact Calvin and rTCA cycles) were detected as well, which might reflect the intensive microbial activities known to occur in this deep biosphere. The presence of those metabolic processes and associated microbes were reflected by functional and genetic markers generated from the metagenomic data in the current study. However, their roles and contributions to the nitrogen/carbon biogeochemical cycles and flux in the abyssal-hadal transition zone still need further analysis. IMPORTANCE The Mariana Trench is the deepest oceanic region on earth, its microbial ecological exploration has become feasible with the rapid progress of submersible and metagenomic sequencing. We investigated the community compositions and metabolic functions of the core microbiome along the abyssal-hadal transition zone of the Mariana Trench, although most studies by far were focused on the pelagic zone. We found a predominance of heterotrophic groups and related metabolic pathways, which were closely associated with nitrogen biogeochemical cycles driven by various nitrifying groups with the capability of dark carbon fixation.
Keywords: Mariana Trench; abyssal-hadal transition zone; core microbiome; metagenomics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

