Development of a Sequence Searchable Database of Celiac Disease-Associated Peptides and Proteins for Risk Assessment of Novel Food Proteins
- PMID: 35769554
- PMCID: PMC9234867
- DOI: 10.3389/falgy.2022.900573
Development of a Sequence Searchable Database of Celiac Disease-Associated Peptides and Proteins for Risk Assessment of Novel Food Proteins
Abstract
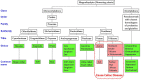
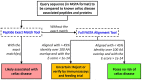
Celiac disease (CeD) is an autoimmune enteropathy induced by prolamin and glutelin proteins in wheat, barley, rye, and triticale recognized by genetically restricted major histocompatibility (MHC) receptors. Patients with CeD must avoid consuming these proteins. Regulators in Europe and the United States expect an evaluation of CeD risks from proteins in genetically modified (GM) crops or novel foods for wheat-related proteins. Our database includes evidence-based causative peptides and proteins and two amino acid sequence comparison tools for CeD risk assessment. Sequence entries are based on the review of published studies of specific gluten-reactive T cell activation or intestinal epithelial toxicity. The initial database in 2012 was updated in 2018 and 2022. The current database holds 1,041 causative peptides and 76 representative proteins. The FASTA sequence comparison of 76 representative CeD proteins provides an insurance for possible unreported epitopes. Validation was conducted using protein homologs from Pooideae and non-Pooideae monocots, dicots, and non-plant proteins. Criteria for minimum percent identity and maximum E-scores are guidelines. Exact matches to any of the 1,041 peptides suggest risks, while FASTA alignment to the 76 CeD proteins suggests possible risks. Matched proteins should be tested further by CeD-specific CD4/8+ T cell assays or in vivo challenges before their use in foods.
Keywords: Pooideae; T-cell epitopes; celiac disease; gluten; peptide database; prolamin; risk assessment; sequence comparison.
Copyright © 2022 Amnuaycheewa, Abdelmoteleb, Wise, Bohle, Ferreira, Tetteh, Taylor and Goodman.
Conflict of interest statement
RG declares limited funding from six biotechnology companies from 2009 to 2012 for support of the database management. Unilever SEAC and NuSeed Americas provided limited funding to the AllergenOnline.org database from 2018 to 2021. These companies did not contribute to or see the article prior to submission. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- van Putten MC, Frewer LJ, Gilissen LJWJ, Gremmen B, Peijnenburg AACM, Wichers HJ. Novel foods and food allergies: a review of the issues. Trends Food Sci Technol. (2006) 17:289 10.1016/j.tifs.2005.11.010 - DOI
-
- Codex Alimentarius Commission . In: Alinorm 03/34: Joint FAO/WHO Food Standard Programme, Codex Alimentarious Commission, Twenty-Fifth Session, Rome, Italy, 30 June−5 July, 2003. Appendix III, Guideline for the conduct of food safety assessment of foods derived from recombinant-DNA plants, and Appendix IV, Annex on the assessment of possible allergenicity. Twenty-Fifth Session (FA); Joint FAO/WHO Food Standards Programme. (2003). p. 47–60.
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

