Identifying SARS-CoV-2 regional introductions and transmission clusters in real time
- PMID: 35769891
- PMCID: PMC9214145
- DOI: 10.1093/ve/veac048
Identifying SARS-CoV-2 regional introductions and transmission clusters in real time
Abstract
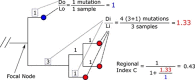
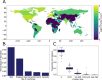
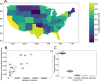
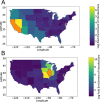
The unprecedented severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) global sequencing effort has suffered from an analytical bottleneck. Many existing methods for phylogenetic analysis are designed for sparse, static datasets and are too computationally expensive to apply to densely sampled, rapidly expanding datasets when results are needed immediately to inform public health action. For example, public health is often concerned with identifying clusters of closely related samples, but the sheer scale of the data prevents manual inspection and the current computational models are often too expensive in time and resources. Even when results are available, intuitive data exploration tools are of critical importance to effective public health interpretation and action. To help address this need, we present a phylogenetic heuristic that quickly and efficiently identifies newly introduced strains in a region, resulting in clusters of infected individuals, and their putative geographic origins. We show that this approach performs well on simulated data and yields results largely congruent with more sophisticated Bayesian phylogeographic modeling approaches. We also introduce Cluster-Tracker (https://clustertracker.gi.ucsc.edu/), a novel interactive web-based tool to facilitate effective and intuitive SARS-CoV-2 geographic data exploration and visualization across the USA. Cluster-Tracker is updated daily and automatically identifies and highlights groups of closely related SARS-CoV-2 infections resulting from the transmission of the virus between two geographic areas by travelers, streamlining public health tracking of local viral diversity and emerging infection clusters. The site is open-source and designed to be easily configured to analyze any chosen region, making it a useful resource globally. The combination of these open-source tools will empower detailed investigations of the geographic origins and spread of SARS-CoV-2 and other densely sampled pathogens.
Keywords: COVID-19; Cluster-Tracker; SARS-CoV-2; genomic epidemiology; phylodynamics; phylogenetic methods; phylogeography.
© The Author(s) 2022. Published by Oxford University Press.
Figures

Similar articles
-
High infectiousness immediately before COVID-19 symptom onset highlights the importance of continued contact tracing.Elife. 2021 Apr 26;10:e65534. doi: 10.7554/eLife.65534. Elife. 2021. PMID: 33899740 Free PMC article.
-
COVID-19 Variant Surveillance and Social Determinants in Central Massachusetts: Development Study.JMIR Form Res. 2022 Jun 13;6(6):e37858. doi: 10.2196/37858. JMIR Form Res. 2022. PMID: 35658093 Free PMC article.
-
The utility of SARS-CoV-2 genomic data for informative clustering under different epidemiological scenarios and sampling.Infect Genet Evol. 2023 Sep;113:105484. doi: 10.1016/j.meegid.2023.105484. Epub 2023 Jul 31. Infect Genet Evol. 2023. PMID: 37531976
-
Bioinformatics resources facilitate understanding and harnessing clinical research of SARS-CoV-2.Brief Bioinform. 2021 Mar 22;22(2):714-725. doi: 10.1093/bib/bbaa416. Brief Bioinform. 2021. PMID: 33432321 Free PMC article. Review.
-
Universal screening for SARS-CoV-2 infection: a rapid review.Cochrane Database Syst Rev. 2020 Sep 15;9(9):CD013718. doi: 10.1002/14651858.CD013718. Cochrane Database Syst Rev. 2020. PMID: 33502003 Free PMC article.
Cited by
-
Genomics-informed outbreak investigations of SARS-CoV-2 using civet.PLOS Glob Public Health. 2022 Dec 9;2(12):e0000704. doi: 10.1371/journal.pgph.0000704. eCollection 2022. PLOS Glob Public Health. 2022. PMID: 36962792 Free PMC article.
-
Variational Phylodynamic Inference Using Pandemic-scale Data.Mol Biol Evol. 2022 Aug 3;39(8):msac154. doi: 10.1093/molbev/msac154. Mol Biol Evol. 2022. PMID: 35816422 Free PMC article.
-
Maximum likelihood pandemic-scale phylogenetics.bioRxiv [Preprint]. 2022 Jul 18:2022.03.22.485312. doi: 10.1101/2022.03.22.485312. bioRxiv. 2022. Update in: Nat Genet. 2023 May;55(5):746-752. doi: 10.1038/s41588-023-01368-0. PMID: 35350209 Free PMC article. Updated. Preprint.
-
SARS-CoV-2 Genomic Epidemiology Dashboards: A Review of Functionality and Technological Frameworks for the Public Health Response.Genes (Basel). 2024 Jul 3;15(7):876. doi: 10.3390/genes15070876. Genes (Basel). 2024. PMID: 39062655 Free PMC article. Review.
-
Optimizing ancestral trait reconstruction of large HIV Subtype C datasets through multiple-trait subsampling.Virus Evol. 2023 Nov 22;9(2):vead069. doi: 10.1093/ve/vead069. eCollection 2023. Virus Evol. 2023. PMID: 38046219 Free PMC article.
References
-
- Brito A. F. et al. (2021) ‘Global Disparities in SARS-CoV-2 Genomic Surveillance’, medRxiv. 2021.08.21.21262393.doi: 10.1101/2021.08.21.21262393. - DOI

