Functional Micropeptides Encoded by Long Non-Coding RNAs: A Comprehensive Review
- PMID: 35769907
- PMCID: PMC9234465
- DOI: 10.3389/fmolb.2022.817517
Functional Micropeptides Encoded by Long Non-Coding RNAs: A Comprehensive Review
Abstract
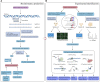
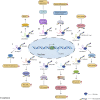
Long non-coding RNAs (lncRNAs) were originally defined as non-coding RNAs (ncRNAs) which lack protein-coding ability. However, with the emergence of technologies such as ribosome profiling sequencing and ribosome-nascent chain complex sequencing, it has been demonstrated that most lncRNAs have short open reading frames hence the potential to encode functional micropeptides. Such micropeptides have been described to be widely involved in life-sustaining activities in several organisms, such as homeostasis regulation, disease, and tumor occurrence, and development, and morphological development of animals, and plants. In this review, we focus on the latest developments in the field of lncRNA-encoded micropeptides, and describe the relevant computational tools and techniques for micropeptide prediction and identification. This review aims to serve as a reference for future research studies on lncRNA-encoded micropeptides.
Keywords: Ribo-seq; coding potential prediction; lncRNA; micropeptide; sORF.
Copyright © 2022 Pan, Wang, Shang, Ma, Rong and Zhang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
Publication types
LinkOut - more resources
Full Text Sources

