Transcriptional Basis for Haustorium Formation and Host Establishment in Hemiparasitic Psittacanthus schiedeanus Mistletoes
- PMID: 35769994
- PMCID: PMC9235361
- DOI: 10.3389/fgene.2022.929490
Transcriptional Basis for Haustorium Formation and Host Establishment in Hemiparasitic Psittacanthus schiedeanus Mistletoes
Abstract
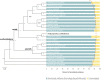
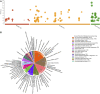
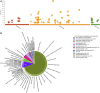
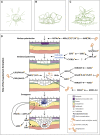
The mistletoe Psittacanthus schiedeanus, a keystone species in interaction networks between plants, pollinators, and seed dispersers, infects a wide range of native and non-native tree species of commercial interest. Here, using RNA-seq methodology we assembled the whole circularized quadripartite structure of P. schiedeanus chloroplast genome and described changes in the gene expression of the nuclear genomes across time of experimentally inoculated seeds. Of the 140,467 assembled and annotated uniGenes, 2,000 were identified as differentially expressed (DEGs) and were classified in six distinct clusters according to their expression profiles. DEGs were also classified in enriched functional categories related to synthesis, signaling, homoeostasis, and response to auxin and jasmonic acid. Since many orthologs are involved in lateral or adventitious root formation in other plant species, we propose that in P. schiedeanus (and perhaps in other rootless mistletoe species), these genes participate in haustorium formation by complex regulatory networks here described. Lastly, and according to the structural similarities of P. schiedeanus enzymes with those that are involved in host cell wall degradation in fungi, we suggest that a similar enzymatic arsenal is secreted extracellularly and used by mistletoes species to easily parasitize and break through tissues of the host.
Keywords: Psittacanthus schiedeanus; haustorium; mistletoe; parasitic plant; transcriptome.
Copyright © 2022 Ibarra-Laclette, Venancio-Rodríguez, Vásquez-Aguilar, Alonso-Sánchez, Pérez-Torres, Villafán, Ramírez-Barahona, Galicia, Sosa, Rebollar, Lara, González-Rodríguez, Díaz-Fleisher and Ornelas.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
LinkOut - more resources
Full Text Sources

