Gene expression profiling of Group 3 medulloblastomas defines a clinically tractable stratification based on KIRREL2 expression
- PMID: 35771282
- PMCID: PMC9288368
- DOI: 10.1007/s00401-022-02460-1
Gene expression profiling of Group 3 medulloblastomas defines a clinically tractable stratification based on KIRREL2 expression
Abstract
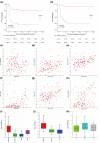
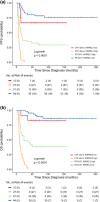
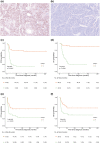
Medulloblastomas (MB) molecularly designated as Group 3 (Grp 3) MB represent a more clinically aggressive tumor variant which, as a group, displays heterogeneous molecular characteristics and disease outcomes. Reliable risk stratification of Grp 3 MB would allow for appropriate assignment of patients to aggressive treatment protocols and, vice versa, for sparing adverse effects of high-dose radio-chemotherapy in patients with standard or low-risk tumors. Here we performed RNA-based analysis on an international cohort of 179 molecularly designated Grp 3 MB treated with HIT protocols. We analyzed the clinical significance of differentially expressed genes, thereby developing optimal prognostic subdivision of this MB molecular group. We compared the transcriptome profiles of two Grp 3 MB subsets with various outcomes (76 died within the first 60 months vs. 103 survived this period) and identified 224 differentially expressed genes (DEG) between these two clinical groups (Limma R algorithm, adjusted p-value < 0.05). We selected the top six DEG overexpressed in the unfavorable cohort for further survival analysis and found that expression of all six genes strongly correlated with poor outcomes. However, only high expression of KIRREL2 was identified as an independent molecular prognostic indicator of poor patients' survival. Based on clinical and molecular patterns, four risk categories were outlined for Grp 3 MB patients: i. low-risk: M0-1/MYC non-amplified/KIRREL2 low (n = 48; 5-year OS-95%); ii. standard-risk: M0-1/MYC non-amplified/KIRREL2 high or M2-3/MYC non-amplified/KIRREL2 low (n = 65; 5-year OS-70%); iii. high-risk: M2-3/MYC non-amplified/KIRREL2 high (n = 36; 5-year OS-30%); iv. very high risk-all MYC amplified tumors (n = 30; 5-year OS-0%). Cross-validated survival models incorporating KIRREL2 expression with clinical features allowed for the reclassification of up to 50% of Grp 3 MB patients into a more appropriate risk category. Finally, KIRREL2 immunopositivity was also identified as a predictive indicator of Grp 3 MB poor survival, thus suggesting its application as a possible prognostic marker in routine clinical settings. Our results indicate that integration of KIRREL2 expression in risk stratification models may improve Grp 3 MB outcome prediction. Therefore, simple gene and/or protein expression analyses for this molecular marker could be easily adopted for Grp 3 MB prognostication and may help in assigning patients to optimal therapeutic approaches in prospective clinical trials.
Keywords: Expression; Group 3; KIRREL2; Medulloblastoma; Prognosis.
© 2022. The Author(s).
Figures


Similar articles
-
Transcriptome analysis stratifies second-generation non-WNT/non-SHH medulloblastoma subgroups into clinically tractable subtypes.Acta Neuropathol. 2023 Jun;145(6):829-842. doi: 10.1007/s00401-023-02575-z. Epub 2023 Apr 24. Acta Neuropathol. 2023. PMID: 37093271
-
Integrated molecular analysis of adult sonic hedgehog (SHH)-activated medulloblastomas reveals two clinically relevant tumor subsets with VEGFA as potent prognostic indicator.Neuro Oncol. 2021 Sep 1;23(9):1576-1585. doi: 10.1093/neuonc/noab031. Neuro Oncol. 2021. PMID: 33589929 Free PMC article.
-
Clinically unfavorable transcriptome subtypes of non-WNT/non-SHH medulloblastomas are associated with a predominance in proliferating and progenitor-like cell subpopulations.Acta Neuropathol. 2024 Jun 7;147(1):95. doi: 10.1007/s00401-024-02746-6. Acta Neuropathol. 2024. PMID: 38847845
-
Molecular Stratification of Medulloblastoma: Clinical Outcomes and Therapeutic Interventions.Anticancer Res. 2022 May;42(5):2225-2239. doi: 10.21873/anticanres.15703. Anticancer Res. 2022. PMID: 35489737 Review.
-
Molecular Classification of Medulloblastoma.Neurol Med Chir (Tokyo). 2016 Nov 15;56(11):687-697. doi: 10.2176/nmc.ra.2016-0016. Epub 2016 May 26. Neurol Med Chir (Tokyo). 2016. PMID: 27238212 Free PMC article. Review.
Cited by
-
Oncogene aberrations drive medulloblastoma progression, not initiation.Nature. 2025 Jun;642(8069):1062-1072. doi: 10.1038/s41586-025-08973-5. Epub 2025 May 7. Nature. 2025. PMID: 40335697 Free PMC article.
-
Genetic Discrimination of Grade 3 and Grade 4 Gliomas by Artificial Neural Network.Cell Mol Neurobiol. 2023 Dec 27;44(1):13. doi: 10.1007/s10571-023-01448-z. Cell Mol Neurobiol. 2023. PMID: 38150033 Free PMC article.
-
Oncolytic virus-driven immune remodeling revealed in mouse medulloblastomas at single cell resolution.Mol Ther Oncolytics. 2023 Jul 19;30:39-55. doi: 10.1016/j.omto.2023.07.006. eCollection 2023 Sep 21. Mol Ther Oncolytics. 2023. PMID: 37583388 Free PMC article.
-
Molecular and clinical heterogeneity within MYC-family amplified medulloblastoma is associated with survival outcomes: A multicenter cohort study.Neuro Oncol. 2025 Jan 12;27(1):222-236. doi: 10.1093/neuonc/noae178. Neuro Oncol. 2025. PMID: 39377358 Free PMC article.
-
Candidate Biomarkers for Targeting in Type 1 Diabetes; A Bioinformatic Analysis of Pancreatic Cell Surface Antigens.Cell J. 2024 Jan 31;26(1):51-61. doi: 10.22074/cellj.2023.1996297.1262. Cell J. 2024. PMID: 38351729 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

