NCOA4-Mediated Ferritinophagy Is a Pancreatic Cancer Dependency via Maintenance of Iron Bioavailability for Iron-Sulfur Cluster Proteins
- PMID: 35771492
- PMCID: PMC9437572
- DOI: 10.1158/2159-8290.CD-22-0043
NCOA4-Mediated Ferritinophagy Is a Pancreatic Cancer Dependency via Maintenance of Iron Bioavailability for Iron-Sulfur Cluster Proteins
Abstract
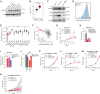
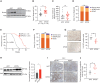
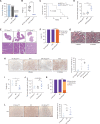
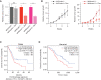
Pancreatic ductal adenocarcinomas (PDAC) depend on autophagy for survival; however, the metabolic substrates that autophagy provides to drive PDAC progression are unclear. Ferritin, the cellular iron storage complex, is targeted for lysosomal degradation (ferritinophagy) by the selective autophagy adaptor NCOA4, resulting in release of iron for cellular utilization. Using patient-derived and murine models of PDAC, we demonstrate that ferritinophagy is upregulated in PDAC to sustain iron availability, thereby promoting tumor progression. Quantitative proteomics reveals that ferritinophagy fuels iron-sulfur cluster protein synthesis to support mitochondrial homeostasis. Targeting NCOA4 leads to tumor growth delay and prolonged survival but with the development of compensatory iron acquisition pathways. Finally, enhanced ferritinophagy accelerates PDAC tumorigenesis, and an elevated ferritinophagy expression signature predicts for poor prognosis in patients with PDAC. Together, our data reveal that the maintenance of iron homeostasis is a critical function of PDAC autophagy, and we define NCOA4-mediated ferritinophagy as a therapeutic target in PDAC.
Significance: Autophagy and iron metabolism are metabolic dependencies in PDAC. However, targeted therapies for these pathways are lacking. We identify NCOA4-mediated selective autophagy of ferritin ("ferritinophagy") as upregulated in PDAC. Ferritinophagy supports PDAC iron metabolism and thereby tumor progression and represents a new therapeutic target in PDAC. See related commentary by Jain and Amaravadi, p. 2023. See related article by Ravichandran et al., p. 2198. This article is highlighted in the In This Issue feature, p. 2007.
©2022 The Authors; Published by the American Association for Cancer Research.
Figures


Comment in
-
Coordinated Transcriptional and Catabolic Programs Support Iron-Dependent Adaptation to RAS-MAPK Pathway Inhibition in Pancreatic Cancer.Cancer Discov. 2022 Sep 2;12(9):2198-2219. doi: 10.1158/2159-8290.CD-22-0044. Cancer Discov. 2022. PMID: 35771494 Free PMC article.
-
Pumping Iron: Ferritinophagy Promotes Survival and Therapy Resistance in Pancreatic Cancer.Cancer Discov. 2022 Sep 2;12(9):2023-2025. doi: 10.1158/2159-8290.CD-22-0734. Cancer Discov. 2022. PMID: 36052501 Free PMC article.
Comment on
-
Coordinated Transcriptional and Catabolic Programs Support Iron-Dependent Adaptation to RAS-MAPK Pathway Inhibition in Pancreatic Cancer.Cancer Discov. 2022 Sep 2;12(9):2198-2219. doi: 10.1158/2159-8290.CD-22-0044. Cancer Discov. 2022. PMID: 35771494 Free PMC article.
References
-
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019 (US statistics). CA Cancer J Clin 2019;69:7–34. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

