Methods to monitor bacterial growth and replicative rates at the single-cell level
- PMID: 35772001
- PMCID: PMC9629498
- DOI: 10.1093/femsre/fuac030
Methods to monitor bacterial growth and replicative rates at the single-cell level
Abstract
The heterogeneity of bacterial growth and replicative rates within a population was proposed a century ago notably to explain the presence of bacterial persisters. The term "growth rate" at the single-cell level corresponds to the increase in size or mass of an individual bacterium while the "replicative rate" refers to its division capacity within a defined temporality. After a decades long hiatus, recent technical innovative approaches allow population growth and replicative rates heterogeneity monitoring at the single-cell level resuming in earnest. Among these techniques, the oldest and widely used is time-lapse microscopy, most recently combined with microfluidics. We also discuss recent fluorescence dilution methods informing only on replicative rates and best suited. Some new elegant single cell methods so far only sporadically used such as buoyant mass measurement and stable isotope probing have emerged. Overall, such tools are widely used to investigate and compare the growth and replicative rates of bacteria displaying drug-persistent behaviors to that of bacteria growing in specific ecological niches or collected from patients. In this review, we describe the current methods available, discussing both the type of queries these have been used to answer and the specific strengths and limitations of each method.
© The Author(s) 2022. Published by Oxford University Press on behalf of FEMS.
Figures

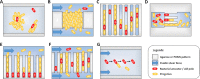
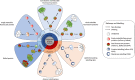
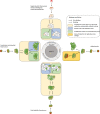

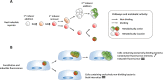


Similar articles
-
Investigating the physiology of viable but non-culturable bacteria by microfluidics and time-lapse microscopy.BMC Biol. 2017 Dec 21;15(1):121. doi: 10.1186/s12915-017-0465-4. BMC Biol. 2017. PMID: 29262826 Free PMC article.
-
Measuring the Replicative Lifespan of Saccharomyces cerevisiae Using the HYAA Microfluidic Platform.Methods Mol Biol. 2020;2144:1-6. doi: 10.1007/978-1-0716-0592-9_1. Methods Mol Biol. 2020. PMID: 32410020 Free PMC article.
-
Single-cell analysis of mycobacteria using microfluidics and time-lapse microscopy.Methods Mol Biol. 2015;1285:241-56. doi: 10.1007/978-1-4939-2450-9_14. Methods Mol Biol. 2015. PMID: 25779320
-
Single-Molecule Insights Into the Dynamics of Replicative Helicases.Front Mol Biosci. 2021 Aug 26;8:741718. doi: 10.3389/fmolb.2021.741718. eCollection 2021. Front Mol Biosci. 2021. PMID: 34513934 Free PMC article. Review.
-
Analyzing Microbial Population Heterogeneity-Expanding the Toolbox of Microfluidic Single-Cell Cultivations.J Mol Biol. 2019 Nov 22;431(23):4569-4588. doi: 10.1016/j.jmb.2019.04.025. Epub 2019 Apr 26. J Mol Biol. 2019. PMID: 31034885 Review.
Cited by
-
Tracking the progeny of bacterial persisters using a CRISPR-based genomic recorder.Proc Natl Acad Sci U S A. 2024 Oct 15;121(42):e2405983121. doi: 10.1073/pnas.2405983121. Epub 2024 Oct 7. Proc Natl Acad Sci U S A. 2024. PMID: 39374386 Free PMC article.
-
Combinatorial fluorescent labeling of live anaerobic bacteria via the incorporation of azide-modified sugars into newly synthesized macromolecules.Nat Protoc. 2023 Dec;18(12):3767-3786. doi: 10.1038/s41596-023-00896-7. Epub 2023 Oct 11. Nat Protoc. 2023. PMID: 37821626 Review.
-
Recent Advances in Bacterial Persistence Mechanisms.Int J Mol Sci. 2023 Sep 20;24(18):14311. doi: 10.3390/ijms241814311. Int J Mol Sci. 2023. PMID: 37762613 Free PMC article. Review.

