Inherited basis of visceral, abdominal subcutaneous and gluteofemoral fat depots
- PMID: 35773277
- PMCID: PMC9247093
- DOI: 10.1038/s41467-022-30931-2
Inherited basis of visceral, abdominal subcutaneous and gluteofemoral fat depots
Abstract
For any given level of overall adiposity, individuals vary considerably in fat distribution. The inherited basis of fat distribution in the general population is not fully understood. Here, we study up to 38,965 UK Biobank participants with MRI-derived visceral (VAT), abdominal subcutaneous (ASAT), and gluteofemoral (GFAT) adipose tissue volumes. Because these fat depot volumes are highly correlated with BMI, we additionally study six local adiposity traits: VAT adjusted for BMI and height (VATadj), ASATadj, GFATadj, VAT/ASAT, VAT/GFAT, and ASAT/GFAT. We identify 250 independent common variants (39 newly-identified) associated with at least one trait, with many associations more pronounced in female participants. Rare variant association studies extend prior evidence for PDE3B as an important modulator of fat distribution. Local adiposity traits (1) highlight depot-specific genetic architecture and (2) enable construction of depot-specific polygenic scores that have divergent associations with type 2 diabetes and coronary artery disease. These results - using MRI-derived, BMI-independent measures of local adiposity - confirm fat distribution as a highly heritable trait with important implications for cardiometabolic health outcomes.
© 2022. The Author(s).
Conflict of interest statement
S.A. has served as a scientific consultant to Third Rock Ventures. M.D.R.K., A.P., and P.B. are supported by grants from Bayer AG applying machine learning in cardiovascular disease. P.T.E. receives sponsored research support from Bayer AG and IBM and has consulted for Bayer AG, Novartis, MyoKardia and Quest Diagnostics. A.P. is also employed as a Venture Partner at GV and consulted for Novartis; and has received funding from Intel, Verily and MSFT. M.C. holds equity in Waypoint Bio and is a member of the Nestle Scientific Advisory Board. K.N. is an employee of IBM Research. P.B. serves as a consultant for Novartis. A.V.K. is an employee and holds equity in Verve Therapeutics; has served as a scientific advisor to Amgen, Maze Therapeutics, Navitor Pharmaceuticals, Sarepta Therapeutics, Novartis, Silence Therapeutics, Korro Bio, Veritas International, Color Health, Third Rock Ventures, Illumina, Foresite Labs, and Columbia University (NIH); received speaking fees from Illumina, MedGenome, Amgen, and the Novartis Institute for Biomedical Research; and received a sponsored research agreement from IBM Research. The remaining authors declare no competing interests.
Figures

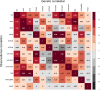


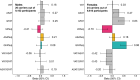


References
-
- González-Muniesa P, et al. Obesity. Nat. Rev. Dis. Prim. 2017;3:1–18. - PubMed
-
- Agrawal, S. et al. Association of machine learning-derived measures of body fat distribution in >40,000 individuals with cardiometabolic diseases. medRxiv. 10.1101/2021.05.07.21256854 (2021).
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

