Frequent aneuploidy in primary human T cells after CRISPR-Cas9 cleavage
- PMID: 35773341
- PMCID: PMC7613940
- DOI: 10.1038/s41587-022-01377-0
Frequent aneuploidy in primary human T cells after CRISPR-Cas9 cleavage
Abstract
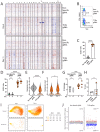
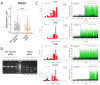
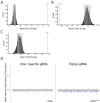
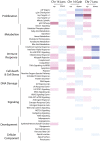
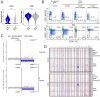
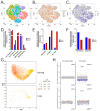
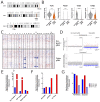
Multiple clinical trials of allogeneic T cell therapy use site-specific nucleases to disrupt T cell receptor (TCR) and other genes1-6. In this study, using single-cell RNA sequencing, we investigated genome editing outcomes in primary human T cells transfected with CRISPR-Cas9 and guide RNAs targeting genes for TCR chains and programmed cell death protein 1. Four days after transfection, we found a loss of chromosome 14, harboring the TCRα locus, in up to 9% of the cells and a chromosome 14 gain in up to 1.4% of the cells. Chromosome 7, harboring the TCRβ locus, was truncated in 9.9% of the cells. Aberrations were validated using fluorescence in situ hybridization and digital droplet PCR. Aneuploidy was associated with reduced proliferation, induced p53 activation and cell death. However, at 11 days after transfection, 0.9% of T cells still had a chromosome 14 loss. Aneuploidy and chromosomal truncations are, thus, frequent outcomes of CRISPR-Cas9 cleavage that should be monitored and minimized in clinical protocols.
© 2022. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
A.B is a co-founder and an inventor of the underlying patents for LogicBio Therapeutics, developing CRISPR-free genome editing.
A.D.N and A.B. are co-founders and inventors of the underlying patents for Tabby Therapeutics, using CRISPR-Cas9 for B cell engineering. A.D.N and A.B hold equity and receive monetary compensation from Tabby Therapeutics. E.R. is an employee of Future Meat Technologies.
Figures




References
-
- Philip LPB, et al. Multiplex genome-edited T-cell manufacturing platform for ‘off-the-shelf’ adoptive T-cell immunotherapies. Cancer Res. 2015;75:3853–3864. - PubMed
-
- Qasim W, et al. Molecular remission of infant B-ALL after infusion of universal TALEN gene-edited CAR T cells. Sci Transl Med. 2017;9:1–10. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

