Telomeric 8-oxo-guanine drives rapid premature senescence in the absence of telomere shortening
- PMID: 35773409
- PMCID: PMC9287163
- DOI: 10.1038/s41594-022-00790-y
Telomeric 8-oxo-guanine drives rapid premature senescence in the absence of telomere shortening
Abstract
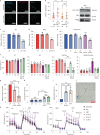
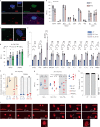
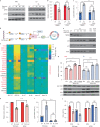
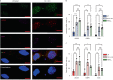
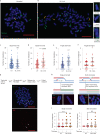
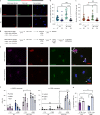
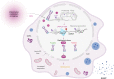
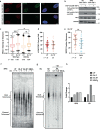
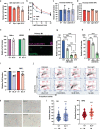
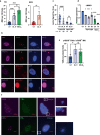
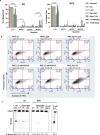
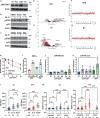
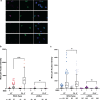
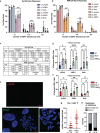
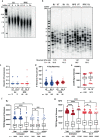
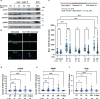
Oxidative stress is a primary cause of cellular senescence and contributes to the etiology of numerous human diseases. Oxidative damage to telomeric DNA has been proposed to cause premature senescence by accelerating telomere shortening. Here, we tested this model directly using a precision chemoptogenetic tool to produce the common lesion 8-oxo-guanine (8oxoG) exclusively at telomeres in human fibroblasts and epithelial cells. A single induction of telomeric 8oxoG is sufficient to trigger multiple hallmarks of p53-dependent senescence. Telomeric 8oxoG activates ATM and ATR signaling, and enriches for markers of telomere dysfunction in replicating, but not quiescent cells. Acute 8oxoG production fails to shorten telomeres, but rather generates fragile sites and mitotic DNA synthesis at telomeres, indicative of impaired replication. Based on our results, we propose that oxidative stress promotes rapid senescence by producing oxidative base lesions that drive replication-dependent telomere fragility and dysfunction in the absence of shortening and shelterin loss.
© 2022. The Author(s).
Conflict of interest statement
M.P.B. is a founder in Sharp Edge Labs, a company applying the FAP-fluorogen technology commercially. The remaining authors declare no competing interests.
Figures
Similar articles
-
OGG1 and MUTYH repair activities promote telomeric 8-oxoguanine induced senescence in human fibroblasts.Nat Commun. 2025 Jan 21;16(1):893. doi: 10.1038/s41467-024-55638-4. Nat Commun. 2025. PMID: 39837827 Free PMC article.
-
Oxidative guanine base damage plays a dual role in regulating productive ALT-associated homology-directed repair.Cell Rep. 2024 Jan 23;43(1):113656. doi: 10.1016/j.celrep.2023.113656. Epub 2024 Jan 8. Cell Rep. 2024. PMID: 38194346 Free PMC article.
-
Oxidative Stress Induces Telomere Dysfunction and Senescence by Replication Fork Arrest.Cells. 2019 Jan 3;8(1):19. doi: 10.3390/cells8010019. Cells. 2019. PMID: 30609792 Free PMC article.
-
Replication Stress at Telomeric and Mitochondrial DNA: Common Origins and Consequences on Ageing.Int J Mol Sci. 2019 Oct 8;20(19):4959. doi: 10.3390/ijms20194959. Int J Mol Sci. 2019. PMID: 31597307 Free PMC article. Review.
-
DNA damage in telomeres and mitochondria during cellular senescence: is there a connection?Nucleic Acids Res. 2007;35(22):7505-13. doi: 10.1093/nar/gkm893. Epub 2007 Nov 5. Nucleic Acids Res. 2007. PMID: 17986462 Free PMC article. Review.
Cited by
-
Mammalian telomeric RNA (TERRA) can be translated to produce valine-arginine and glycine-leucine dipeptide repeat proteins.Proc Natl Acad Sci U S A. 2023 Feb 28;120(9):e2221529120. doi: 10.1073/pnas.2221529120. Epub 2023 Feb 22. Proc Natl Acad Sci U S A. 2023. PMID: 36812212 Free PMC article.
-
Arsenite exposure induces premature senescence and senescence-associated secretory phenotype (SASP) in human hepatocyte-derived cell line Huh-7.Environ Health Prev Med. 2024;29:74. doi: 10.1265/ehpm.24-00139. Environ Health Prev Med. 2024. PMID: 39756915 Free PMC article.
-
Distinct regulation of ATM signaling by DNA single-strand breaks and APE1.Nat Commun. 2024 Aug 7;15(1):6517. doi: 10.1038/s41467-024-50836-6. Nat Commun. 2024. PMID: 39112456 Free PMC article.
-
Cellular Senescence in Acute Liver Injury: What Happens to the Young Liver?Aging Dis. 2024 Jun 10;16(3):1347-1362. doi: 10.14336/AD.2024.0586. Aging Dis. 2024. PMID: 38913043 Free PMC article. Review.
-
New hallmarks of ageing: a 2022 Copenhagen ageing meeting summary.Aging (Albany NY). 2022 Aug 29;14(16):6829-6839. doi: 10.18632/aging.204248. Epub 2022 Aug 29. Aging (Albany NY). 2022. PMID: 36040386 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

