Emu: species-level microbial community profiling of full-length 16S rRNA Oxford Nanopore sequencing data
- PMID: 35773532
- PMCID: PMC9939874
- DOI: 10.1038/s41592-022-01520-4
Emu: species-level microbial community profiling of full-length 16S rRNA Oxford Nanopore sequencing data
Abstract
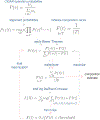
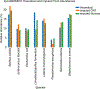
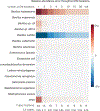
16S ribosomal RNA-based analysis is the established standard for elucidating the composition of microbial communities. While short-read 16S rRNA analyses are largely confined to genus-level resolution at best, given that only a portion of the gene is sequenced, full-length 16S rRNA gene amplicon sequences have the potential to provide species-level accuracy. However, existing taxonomic identification algorithms are not optimized for the increased read length and error rate often observed in long-read data. Here we present Emu, an approach that uses an expectation-maximization algorithm to generate taxonomic abundance profiles from full-length 16S rRNA reads. Results produced from simulated datasets and mock communities show that Emu is capable of accurate microbial community profiling while obtaining fewer false positives and false negatives than alternative methods. Additionally, we illustrate a real-world application of Emu by comparing clinical sample composition estimates generated by an established whole-genome shotgun sequencing workflow with those returned by full-length 16S rRNA gene sequences processed with Emu.
© 2022. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
Competing Interests Statement
The authors declare no competing interests.
Figures




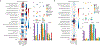

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

