Metabarcoding of Fish Larvae in the Merbok River Reveals Species Diversity and Distribution Along its Mangrove Environment
- PMID: 35774258
- PMCID: PMC9169113
- DOI: 10.6620/ZS.2021.60-76
Metabarcoding of Fish Larvae in the Merbok River Reveals Species Diversity and Distribution Along its Mangrove Environment
Abstract
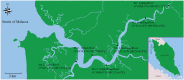
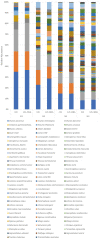
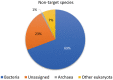
The Merbok River (north-west of Peninsular Malaysia) is a mangrove estuary that provides habitat for over 100 species of fish, which are economically and ecologically important. Threats such as habitat loss and overfishing are becoming a great concern for fisheries conservation and management. The identification of larval fish in this estuarine system is important to complement information on the adults. This is because the data could inform the spawning behaviour, reproductive biology, selection of nursery grounds and migration route of fish. Such information is invaluable for fisheries and aquatic environmental monitoring, and thus for their conservation and management. However, identifying fish larvae is a challenging task based only on morphology and even traditional DNA barcoding. To address this, DNA metabarcoding was utilised to detect the diversity of fish in the Merbok River. To complete the study, the fish larvae were collected at six sampling sites of the river. The extracted larval DNA was amplified for the Cytochrome Oxidase subunit 1 (COI) and 12S ribosomal RNA (12S rRNA) genes based on the metabarcoding approach using shotgun sequencing on the next-generation sequencing (NGS) Illumina MiSeq platform. Eighty-nine species from 65 genera and 41 families were detected, with Oryzias javanicus, Oryzias dancena, Lutjanus argentimaculatus and Lutjanus malabaricus among the most common species. The lower diversity observed from previous morphological studies is suggested to be mainly due to seasonal variation over the sampling period between the two methods and limited 12S rRNA sequences in current databases. The metabarcode data and a validation Sanger sequencing step using 15 species-specific primer pairs detected three species in common: Oryzias javanicus, Decapterus maruadsi and Pennahia macrocephalus. Several discrepancies observed between the two molecular approaches could be attributed to contaminants during sampling and DNA extraction, which could mask the presence of target species, especially when DNA from the contaminants is more abundant than the target organisms. In conclusion, this rapid and cost-effective identification method using DNA metabarcoding allowed the detection of numerous fish species from bulk larval samples in the Merbok River. This method can be applied to other sites and other organisms of interest.
Keywords: DNA metabarcoding; Fish larvae; Mangrove estuary; Merbok River; Next-generation sequencing.
Figures

References
-
- Ambak MA, Mansor MI, Zakaria MZ, Mazlan AG. 2012. Fishes of Malaysia, Univeristi Malaysia Terengganu, Kuala Terengganu, Terengganu, Penerbit UMT.
-
- Arshad AB, Ara R, Amin SMN, Daud SK, Ghaffar MA. 2012. Larval fish composition and spatio-temporal variation in the estuary of Pendas River, southwestern Johor, Peninsular Malaysia. Coastal Marine Science 35(1):96‒102.
-
- Atan Y, Jaafar H, Majid ARA. 2010. Ikan Laut Malaysia: glosari nama sahih spesies ikan, Dewan Bahasa dan Pustaka.
-
- Azmir I, Esa Y, Amin S, Md Yasin I, Md Yusof F. 2017. Identification of larval fish in mangrove areas of Peninsular Malaysia using morphology and DNA barcoding methods. J Appl Ichthyol 33(5):998‒1006. doi:10.1111/jai.13425.
LinkOut - more resources
Full Text Sources
