Differentially Expressed Bone Marrow microRNAs Are Associated With Soluble HLA-G Bone Marrow Levels in Childhood Leukemia
- PMID: 35774498
- PMCID: PMC9237524
- DOI: 10.3389/fgene.2022.871972
Differentially Expressed Bone Marrow microRNAs Are Associated With Soluble HLA-G Bone Marrow Levels in Childhood Leukemia
Abstract
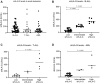
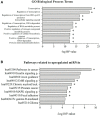
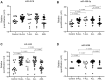
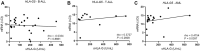
HLA-G is a nonclassical histocompatibility class I molecule that plays a role in immune vigilance in cancer and infectious diseases. We previously reported that highly soluble HLA-G (sHLA-G) levels in the bone marrow were associated with a high blood cell count in T-acute lymphoblastic leukemia, a marker associated with a poor prognosis. To understand the posttranscriptional HLA-G gene regulation in leukemia, we evaluated the bone marrow microRNA profile associated with the HLA-G bone marrow mRNA expression and sHLA-G bone marrow levels in children exhibiting acute leukemia (B-ALL, T-ALL, and AML) using massively parallel sequencing. Ten differentially expressed miRNAs were associated with high sHLA-G bone marrow levels, and four of them (hsa-miR-4516, hsa-miR-486-5p, hsa-miR-4488, and hsa-miR-5096) targeted HLA-G, acting at distinct HLA-G gene segments. For qPCR validation, these miRNA expression levels (ΔCt) were correlated with HLA-G5 and RREB1 mRNA expressions and sHLA-G bone marrow levels according to the leukemia subtype. The hsa-miR-4488 and hsa-miR-5096 expression levels were lower in B-ALL than in AML, while that of hsa-miR-486-5p was lower in T-ALL than in AML. In T-ALL, hsa-miR-5096 correlated positively with HLA-G5 and negatively with sHLA-G. In addition, hsa-miR-4516 correlated negatively with sHLA-G levels. In AML, hsa-miR-4516 and hsa-miR-4488 correlated positively with HLA-G5 mRNA, but the HLA-G5 negatively correlated with sHLA-G. Our findings highlight the need to validate the findings of massively parallel sequencing since the experiment generally uses few individuals, and the same type of leukemia can be molecularly quite variable. We showed that miRNA's milieu in leukemia's bone marrow environment varies according to the type of leukemia and that the regulation of sHLA-G expression exerted by the same miRNA may act by a distinct mechanism in different types of leukemia.
Keywords: ALL; AML; HLA-G; bone marrow; leukemia; microRNA; posttranscriptional regulation.
Copyright © 2022 Almeida, Gomes, Araújo, Oliveira, Santos, Donadi and Lucena-Silva.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
-
- Almeida R. S., Araújo F. S., Oliveira S. A. V., Coutinho L. L., Donadi E. A., Lucena‐Silva N. (2018). “Mirnome Analysis of Brazilian Childhood Acute Lymphoblastic Leukemia Reveals an Association of 10 miRNAs with Increased Soluble HLA-G Levels,” in Abstracts from the 50th Congress of the International Society of Paediatric Oncology (SIOP) Kyoto, Japan November 16-19, 2018 (Pediatr: Blood Cancer; ). 10.1002/pbc.27455 - DOI
LinkOut - more resources
Full Text Sources
Research Materials

