PhyloHerb: A high-throughput phylogenomic pipeline for processing genome skimming data
- PMID: 35774988
- PMCID: PMC9215275
- DOI: 10.1002/aps3.11475
PhyloHerb: A high-throughput phylogenomic pipeline for processing genome skimming data
Abstract
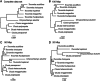
Premise: The application of high-throughput sequencing, especially to herbarium specimens, is rapidly accelerating biodiversity research. Low-coverage sequencing of total genomic DNA (genome skimming) is particularly promising and can simultaneously recover the plastid, mitochondrial, and nuclear ribosomal regions across hundreds of species. Here, we introduce PhyloHerb, a bioinformatic pipeline to efficiently assemble phylogenomic data sets derived from genome skimming.
Methods and results: PhyloHerb uses either a built-in database or user-specified references to extract orthologous sequences from all three genomes using a BLAST search. It outputs FASTA files and offers a suite of utility functions to assist with alignment, partitioning, concatenation, and phylogeny inference. The program is freely available at https://github.com/lmcai/PhyloHerb/.
Conclusions: We demonstrate that PhyloHerb can accurately identify genes using a published data set from Clusiaceae. We also show via simulations that our approach is effective for highly fragmented assemblies from herbarium specimens and is scalable to thousands of species.
Keywords: herbariomics; high‐throughput sequencing; mitochondria; plastome; ribosomal genes.
© 2022 The Authors. Applications in Plant Sciences published by Wiley Periodicals LLC on behalf of Botanical Society of America.
Figures


References
-
- Bakker, F. T. , Lei D., Yu J., Mohammadin S., Wei Z., van de Kerke S., Gravendeel B., et al. 2016. Herbarium genomics: Plastome sequence assembly from a range of herbarium specimens using an Iterative Organelle Genome Assembly pipeline. Biological Journal of the Linnean Society 117: 33–43.
-
- Cai, L. , Zhang H., and Davis C. C.. 2021. Herbariomics‐based biodiversity research: from specimen to phylogeny. Botany 2021: Annual Meeting of the Botanical Society of America, held online [online abstract]. Website: https://2021.botanyconference.org/engine/search/index.php?func=detail%26... [accessed 19 April 2022].
-
- Doyle, J. J. 2022. Defining coalescent genes: Theory meets practice in organelle phylogenomics. Systematic Biology 71: 476–489. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

