A combinatorial system to examine the enzymatic repair of multiply damaged DNA substrates
- PMID: 35776119
- PMCID: PMC9303388
- DOI: 10.1093/nar/gkac530
A combinatorial system to examine the enzymatic repair of multiply damaged DNA substrates
Abstract
DNA damage drives genetic mutations that underlie the development of cancer in humans. Multiple pathways have been described in mammalian cells which can repair this damage. However, most work to date has focused upon single lesions in DNA. We present here a combinatorial system which allows assembly of duplexes containing single or multiple types of damage by ligating together six oligonucleotides containing damaged or modified bases. The combinatorial system has dual fluorescent labels allowing examination of both strands simultaneously, in order to study interactions or competition between different DNA repair pathways. Using this system, we demonstrate how repair of oxidative damage in one DNA strand can convert a mispaired T:G deamination intermediate into a T:A mutation. We also demonstrate that slow repair of a T:G mispair, relative to a U:G mispair, by the human methyl-binding domain 4 DNA glycosylase provides a competitive advantage to competing repair pathways, and could explain why CpG dinucleotides are hotspots for C to T mutations in human tumors. Data is also presented that suggests repair of closely spaced lesions in opposing strands can be repaired by a combination of short and long-patch base excision repair and simultaneous repair of multiply damage sites can potentially lead to lethal double strand breaks.
© The Author(s) 2022. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

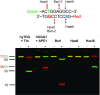







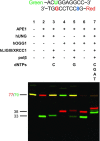
References
-
- Mullaart E., Lohman P.H.M., Berends F., Vijg J.. DNA damage metabolism and aging. Mutat. Res. 1990; 237:189–210. - PubMed
-
- Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993; 362:709–715. - PubMed
-
- Geacintov N.E., Swenberg C.E.. Chemical, molecular biology, and genetic techniques for correlating DNA base damage induced by ionizing radiation with biological end points. Basic Life. Sci. 1991; 58:453–474. - PubMed
-
- Guengerich F.P., Kim M.-S., Müller M., Lowe L.G.. Müller-Hermelink H.K., Neumann H-.G., Dekant W.. Chemical mechanisms of formation of DNA-Carcinogen adducts, elucidation of potential of adducts for mutagenicity, and mechanisms of polymerase fidelity and mutation in the presence of adducts. 1997; Berlin, Heidelberg: Springer Berlin Heidelberg; 49–63.Risk and Progression Factors in Carcinogenesis. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

