A structured model and likelihood approach to estimate yeast prion propagon replication rates and their asymmetric transmission
- PMID: 35776712
- PMCID: PMC9249220
- DOI: 10.1371/journal.pcbi.1010107
A structured model and likelihood approach to estimate yeast prion propagon replication rates and their asymmetric transmission
Abstract
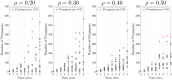
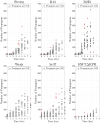
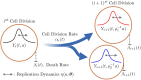
Prion proteins cause a variety of fatal neurodegenerative diseases in mammals but are generally harmless to Baker's yeast (Saccharomyces cerevisiae). This makes yeast an ideal model organism for investigating the protein dynamics associated with these diseases. The rate of disease onset is related to both the replication and transmission kinetics of propagons, the transmissible agents of prion diseases. Determining the kinetic parameters of propagon replication in yeast is complicated because the number of propagons in an individual cell depends on the intracellular replication dynamics and the asymmetric division of yeast cells within a growing yeast cell colony. We present a structured population model describing the distribution and replication of prion propagons in an actively dividing population of yeast cells. We then develop a likelihood approach for estimating the propagon replication rate and their transmission bias during cell division. We first demonstrate our ability to correctly recover known kinetic parameters from simulated data, then we apply our likelihood approach to estimate the kinetic parameters for six yeast prion variants using propagon recovery data. We find that, under our modeling framework, all variants are best described by a model with an asymmetric transmission bias. This demonstrates the strength of our framework over previous formulations assuming equal partitioning of intracellular constituents during cell division.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
The number and transmission of [PSI] prion seeds (Propagons) in the yeast Saccharomyces cerevisiae.PLoS One. 2009;4(3):e4670. doi: 10.1371/journal.pone.0004670. Epub 2009 Mar 5. PLoS One. 2009. PMID: 19262693 Free PMC article.
-
Analysis of the generation and segregation of propagons: entities that propagate the [PSI+] prion in yeast.Genetics. 2003 Sep;165(1):23-33. doi: 10.1093/genetics/165.1.23. Genetics. 2003. PMID: 14504215 Free PMC article.
-
The actin cytoskeletal network plays a role in yeast prion transmission and contributes to prion stability.Mol Microbiol. 2020 Sep;114(3):480-494. doi: 10.1111/mmi.14528. Epub 2020 Jun 8. Mol Microbiol. 2020. PMID: 32426863 Free PMC article.
-
[Yeast as a model for studying the prion and amyloid occurrence].Ross Fiziol Zh Im I M Sechenova. 2004 May;90(5):645-57. Ross Fiziol Zh Im I M Sechenova. 2004. PMID: 15341089 Review. Russian.
-
Impact of Amyloid Polymorphism on Prion-Chaperone Interactions in Yeast.Viruses. 2019 Apr 16;11(4):349. doi: 10.3390/v11040349. Viruses. 2019. PMID: 30995727 Free PMC article. Review.
References
-
- Association A. 2019 Alzheimer’s disease facts and figures. Alzheimer’s & dementia. 2019;15(3):321–387. doi: 10.1016/j.jalz.2019.01.010 - DOI
-
- Head MW. Human prion diseases. Cambridge University Press; 2014.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

