A comprehensive resource for Bordetella genomic epidemiology and biodiversity studies
- PMID: 35778384
- PMCID: PMC9249784
- DOI: 10.1038/s41467-022-31517-8
A comprehensive resource for Bordetella genomic epidemiology and biodiversity studies
Abstract
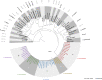
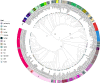
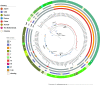
The genus Bordetella includes bacteria that are found in the environment and/or associated with humans and other animals. A few closely related species, including Bordetella pertussis, are human pathogens that cause diseases such as whooping cough. Here, we present a large database of Bordetella isolates and genomes and develop genotyping systems for the genus and for the B. pertussis clade. To generate the database, we merge previously existing databases from Oxford University and Institut Pasteur, import genomes from public repositories, and add 83 newly sequenced B. bronchiseptica genomes. The public database currently includes 2582 Bordetella isolates and their provenance data, and 2085 genomes ( https://bigsdb.pasteur.fr/bordetella/ ). We use core-genome multilocus sequence typing (cgMLST) to develop genotyping systems for the whole genus and for B. pertussis, as well as specific schemes to define antigenic, virulence and macrolide resistance profiles. Phylogenetic analyses allow us to redefine evolutionary relationships among known Bordetella species, and to propose potential new species. Our database provides an expandable resource for genotyping of environmental and clinical Bordetella isolates, thus facilitating evolutionary and epidemiological research on whooping cough and other Bordetella infections.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

