Labeling of a mutant estrogen receptor with an Affimer in a breast cancer cell line
- PMID: 35778844
- PMCID: PMC9617163
- DOI: 10.1016/j.bpj.2022.06.028
Labeling of a mutant estrogen receptor with an Affimer in a breast cancer cell line
Abstract
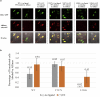
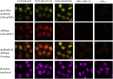
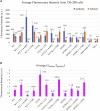
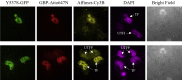
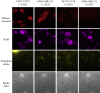
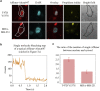
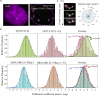
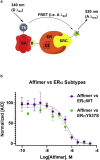
Mutations of the intracellular estrogen receptor alpha (ERα) is implicated in 70% of breast cancers. Therefore, it is of considerable interest to image various mutants (L536S, Y537S, D538G) in living cancer cell lines, particularly as a function of various anticancer drugs. We therefore developed a small (13 kDa) Affimer, which, after fluorescent labeling, is able to efficiently label ERα by traveling through temporary pores in the cell membrane, created by the toxin streptolysin O. The Affimer, selected by a phage display, predominantly labels the Y537S mutant and can tell the difference between L536S and D538G mutants. The vast majority of Affimer-ERαY537S is in the nucleus and is capable of an efficient, unrestricted navigation to its target DNA sequence, as visualized by single-molecule fluorescence. The Affimer can also differentiate the effect of selective estrogen receptor modulators. More generally, this is an example of a small binding reagent-an Affimer protein-that can be inserted into living cells with minimal perturbation and high efficiency, to image an endogenous protein.
Copyright © 2022 Biophysical Society. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures
Similar articles
-
Lasofoxifene as a potential treatment for therapy-resistant ER-positive metastatic breast cancer.Breast Cancer Res. 2021 May 12;23(1):54. doi: 10.1186/s13058-021-01431-w. Breast Cancer Res. 2021. PMID: 33980285 Free PMC article.
-
Stereospecific lasofoxifene derivatives reveal the interplay between estrogen receptor alpha stability and antagonistic activity in ESR1 mutant breast cancer cells.Elife. 2022 May 16;11:e72512. doi: 10.7554/eLife.72512. Elife. 2022. PMID: 35575456 Free PMC article.
-
Cadmium activation of wild-type and constitutively active estrogen receptor alpha.Front Endocrinol (Lausanne). 2024 Aug 9;15:1380047. doi: 10.3389/fendo.2024.1380047. eCollection 2024. Front Endocrinol (Lausanne). 2024. PMID: 39184142 Free PMC article.
-
Strategies to degrade estrogen receptor α in primary and ESR1 mutant-expressing metastatic breast cancer.Mol Cell Endocrinol. 2019 Jan 15;480:107-121. doi: 10.1016/j.mce.2018.10.020. Epub 2018 Oct 31. Mol Cell Endocrinol. 2019. PMID: 30389467 Review.
-
A Closer Look at Estrogen Receptor Mutations in Breast Cancer and Their Implications for Estrogen and Antiestrogen Responses.Int J Mol Sci. 2021 Jan 13;22(2):756. doi: 10.3390/ijms22020756. Int J Mol Sci. 2021. PMID: 33451133 Free PMC article. Review.
Cited by
-
A Stronger IMPACT on Career Development for Early- and Mid-career Faculty.J Endocr Soc. 2024 Nov 14;8(12):bvae191. doi: 10.1210/jendso/bvae191. eCollection 2024 Oct 29. J Endocr Soc. 2024. PMID: 39564580 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

