Editing melon eIF4E associates with virus resistance and male sterility
- PMID: 35778883
- PMCID: PMC9491454
- DOI: 10.1111/pbi.13885
Editing melon eIF4E associates with virus resistance and male sterility
Abstract
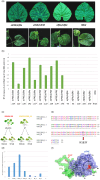
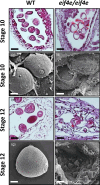
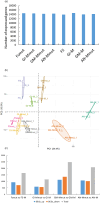
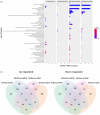
The cap-binding protein eIF4E, through its interaction with eIF4G, constitutes the core of the eIF4F complex, which plays a key role in the circularization of mRNAs and their subsequent cap-dependent translation. In addition to its fundamental role in mRNA translation initiation, other functions have been described or suggested for eIF4E, including acting as a proviral factor and participating in sexual development. We used CRISPR/Cas9 genome editing to generate melon eif4e knockout mutant lines. Editing worked efficiently in melon, as we obtained transformed plants with a single-nucleotide deletion in homozygosis in the first eIF4E exon already in a T0 generation. Edited and non-transgenic plants of a segregating F2 generation were inoculated with Moroccan watermelon mosaic virus (MWMV); homozygous mutant plants showed virus resistance, while heterozygous and non-mutant plants were infected, in agreement with our previous results with plants silenced in eIF4E. Interestingly, all homozygous edited plants of the T0 and F2 generations showed a male sterility phenotype, while crossing with wild-type plants restored fertility, displaying a perfect correlation between the segregation of the male sterility phenotype and the segregation of the eif4e mutation. Morphological comparative analysis of melon male flowers along consecutive developmental stages showed postmeiotic abnormal development for both microsporocytes and tapetum, with clear differences in the timing of tapetum degradation in the mutant versus wild-type. An RNA-Seq analysis identified critical genes in pollen development that were down-regulated in flowers of eif4e/eif4e plants, and suggested that eIF4E-specific mRNA translation initiation is a limiting factor for male gametes formation in melon.
Keywords: cucurbit; pollen; potyvirus resistance; resistance breaking; sexual development; translation initiation.
© 2022 The Authors. Plant Biotechnology Journal published by Society for Experimental Biology and The Association of Applied Biologists and John Wiley & Sons Ltd.
Conflict of interest statement
Authors declare no conflict of interest.
Figures



Similar articles
-
Melon RNA interference (RNAi) lines silenced for Cm-eIF4E show broad virus resistance.Mol Plant Pathol. 2012 Sep;13(7):755-63. doi: 10.1111/j.1364-3703.2012.00785.x. Epub 2012 Feb 6. Mol Plant Pathol. 2012. PMID: 22309030 Free PMC article.
-
Editing eIF4E in the Watermelon Genome Using CRISPR/Cas9 Technology Confers Resistance to ZYMV.Int J Mol Sci. 2024 Oct 25;25(21):11468. doi: 10.3390/ijms252111468. Int J Mol Sci. 2024. PMID: 39519021 Free PMC article.
-
Coordinated and selective recruitment of eIF4E and eIF4G factors for potyvirus infection in Arabidopsis thaliana.FEBS Lett. 2007 Mar 6;581(5):1041-6. doi: 10.1016/j.febslet.2007.02.007. Epub 2007 Feb 14. FEBS Lett. 2007. PMID: 17316629
-
Manipulation of the host translation initiation complex eIF4F by DNA viruses.Biochem Soc Trans. 2010 Dec;38(6):1511-6. doi: 10.1042/BST0381511. Biochem Soc Trans. 2010. PMID: 21118117 Review.
-
eIF4E Resistance: Natural Variation Should Guide Gene Editing.Trends Plant Sci. 2017 May;22(5):411-419. doi: 10.1016/j.tplants.2017.01.008. Epub 2017 Feb 28. Trends Plant Sci. 2017. PMID: 28258958 Review.
Cited by
-
Oligomeric SWEET1g of Solanum tuberosum confers resistance to potato virus Y and Potato virus X.Plant J. 2025 Aug;123(3):e70400. doi: 10.1111/tpj.70400. Plant J. 2025. PMID: 40758984 Free PMC article.
-
Genic male and female sterility in vegetable crops.Hortic Res. 2022 Nov 19;10(1):uhac232. doi: 10.1093/hr/uhac232. eCollection 2023. Hortic Res. 2022. PMID: 36643746 Free PMC article.
-
Validation of CRISPR construct activity and gene function in melon via a hairy root transformation system.Physiol Mol Biol Plants. 2025 May;31(5):753-766. doi: 10.1007/s12298-025-01607-0. Epub 2025 Jun 9. Physiol Mol Biol Plants. 2025. PMID: 40568484
-
State-of-the-Art Molecular Plant Biology Research in Spain.Int J Mol Sci. 2023 Nov 21;24(23):16557. doi: 10.3390/ijms242316557. Int J Mol Sci. 2023. PMID: 38068878 Free PMC article.
-
Rice Yellow Mottle Virus resistance by genome editing of the Oryza sativa L. ssp. japonica nucleoporin gene OsCPR5.1 but not OsCPR5.2.Plant Biotechnol J. 2024 May;22(5):1299-1311. doi: 10.1111/pbi.14266. Epub 2023 Dec 20. Plant Biotechnol J. 2024. PMID: 38124291 Free PMC article.
References
-
- Ariizumi, T. , Hatakeyama, K. , Hinata, K. , Inatsugi, R. , Nishida, I. , Sato, S. , Kato, T. et al. (2004) Disruption of the novel plant protein NEF1 affects lipid accumulation in the plastids of the tapetum and exine formation of pollen, resulting in male sterility in Arabidopsis thaliana. Plant J. 39, 170–181. - PubMed
-
- Ayme, V. , Petit‐Pierre, J. , Souche, S. , Palloix, A. and Moury, B. (2007) Molecular dissection of the potato virus Y VPg virulence factor reveals complex adaptations to the pvr2 resistance allelic series in pepper. J. Gen. Virol. 88, 1594–1601. - PubMed
-
- Bai, S.L. , Peng, Y.B. , Cui, J.X. , Gu, H.T. , Xu, L.Y. , Li, Y.Q. , Xu, Z.H. et al. (2004) Developmental analyses reveal early arrests of the spore‐bearing parts of reproductive organs in unisexual flowers of cucumber (Cucumis sativus L.). Planta 220, 230–240. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Miscellaneous

