Warthog Genomes Resolve an Evolutionary Conundrum and Reveal Introgression of Disease Resistance Genes
- PMID: 35779009
- PMCID: PMC9250280
- DOI: 10.1093/molbev/msac134
Warthog Genomes Resolve an Evolutionary Conundrum and Reveal Introgression of Disease Resistance Genes
Abstract
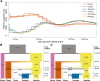
African wild pigs have a contentious evolutionary and biogeographic history. Until recently, desert warthog (Phacochoerus aethiopicus) and common warthog (P. africanus) were considered a single species. Molecular evidence surprisingly suggested they diverged at least 4.4 million years ago, and possibly outside of Africa. We sequenced the first whole-genomes of four desert warthogs and 35 common warthogs from throughout their range. We show that these two species diverged much later than previously estimated, 400,000-1,700,000 years ago depending on assumptions of gene flow. This brings it into agreement with the paleontological record. We found that the common warthog originated in western Africa and subsequently colonized eastern and southern Africa. During this range expansion, the common warthog interbred with the desert warthog, presumably in eastern Africa, underlining this region's importance in African biogeography. We found that immune system-related genes may have adaptively introgressed into common warthogs, indicating that resistance to novel diseases was one of the most potent drivers of evolution as common warthogs expanded their range. Hence, we solve some of the key controversies surrounding warthog evolution and reveal a complex evolutionary history involving range expansion, introgression, and adaptation to new diseases.
Keywords: Phacochoerus evolution; African phylogeography; disease resistance; introgression; population structure.
© The Author(s) 2022. Published by Oxford University Press on behalf of Society for Molecular Biology and Evolution.
Figures




References
-
- FAO . 2012. Global Ecological Zones for FAO Forest Reporting: 2010 Update. Rome, Italy: Food and Agriculture Organization of the United Nations.
-
- Andrews S. 2010. FastQC: a quality control tool for high throughput sequence data [cited 2022 Jun 21]. Available from: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
-
- Antón SC, Potts R, Aiello LC. 2014. Evolution of early Homo: an integrated biological perspective. Science. 345:1236828. - PubMed
-
- Arctander P, Johansen C, Coutellec-Vreto MA. 1999. Phylogeography of three closely related African bovids (tribe Alcelaphini). Mol Biol Evol. 16:1724–1739. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

