Extraction of CRISPR-targeted sequences from the metagenome
- PMID: 35780428
- PMCID: PMC9256941
- DOI: 10.1016/j.xpro.2022.101525
Extraction of CRISPR-targeted sequences from the metagenome
Abstract
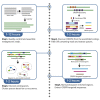
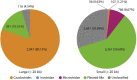
Homology-based search is commonly used to uncover mobile genetic elements (MGEs) from metagenomes, but it heavily relies on reference genomes in the database. Here we introduce a protocol to extract CRISPR-targeted sequences from the assembled human gut metagenomic sequences without using a reference database. We describe the assembling of metagenome contigs, the extraction of CRISPR direct repeats and spacers, the discovery of protospacers, and the extraction of protospacer-enriched regions using the graph-based approach. This protocol could extract numerous characterized/uncharacterized MGEs. For complete details on the use and execution of this protocol, please refer to Sugimoto et al. (2021).
Keywords: Bioinformatics; CRISPR; Genomics; Sequence analysis; Systems biology.
Copyright © 2022 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures
References
-
- Bankevich A., Nurk S., Antipov D., Gurevich A.A., Dvorkin M., Kulikov A.S., Lesin V.M., Nikolenko S.I., Pham S., Prjibelski A.D., et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. - DOI - PMC - PubMed
-
- Bushnell B. BBTools software package. 2014. http://sourceforge. net/projects/bbmap
-
- Cock P.J.A., Antao T., Chang J.T., Chapman B.A., Cox C.J., Dalke A., Friedberg I., Hamelryck T., Kauff F., Wilczynski B., de Hoon M.J.L. Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics. 2009;25:1422–1423. doi: 10.1093/bioinformatics/btp163. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

