Differential Trafficking and Expression of PIR Proteins in Acute and Chronic Plasmodium Infections
- PMID: 35782145
- PMCID: PMC9245118
- DOI: 10.3389/fcimb.2022.877253
Differential Trafficking and Expression of PIR Proteins in Acute and Chronic Plasmodium Infections
Abstract
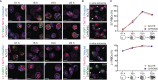
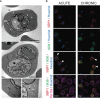
Plasmodium multigene families are thought to play important roles in the pathogenesis of malaria. Plasmodium interspersed repeat (pir) genes comprise the largest multigene family in many Plasmodium species. However, their expression pattern and localisation remain to be elucidated. Understanding protein subcellular localisation is fundamental to reveal the functional importance and cell-cell interactions of the PIR proteins. Here, we use the rodent malaria parasite, Plasmodium chabaudi chabaudi, as a model to investigate the localisation pattern of this gene family. We found that most PIR proteins are co-expressed in clusters during acute and chronic infection; members of the S7 clade are predominantly expressed during the acute-phase, whereas members of the L1 clade dominate the chronic-phase of infection. Using peptide antisera specific for S7 or L1 PIRS, we show that these PIRs have different localisations within the infected red blood cells. S7 PIRs are exported into the infected red blood cell cytoplasm where they are co-localised with parasite-induced host cell modifications termed Maurer's clefts, whereas L1 PIRs are localised on or close to the parasitophorous vacuolar membrane. This localisation pattern changes following mosquito transmission and during progression from acute- to chronic-phase of infection. The presence of PIRs in Maurer's clefts, as seen for Plasmodium falciparum RIFIN and STEVOR proteins, might suggest trafficking of the PIRs on the surface of the infected erythrocytes. However, neither S7 nor L1 PIR proteins detected by the peptide antisera are localised on the surface of infected red blood cells, suggesting that they are unlikely to be targets of surface variant-specific antibodies or to be directly involved in adhesion of infected red blood cells to host cells, as described for Plasmodium falciparum VAR proteins. The differences in subcellular localisation of the two major clades of Plasmodium chabaudi PIRs across the blood cycle, and the apparent lack of expression on the red cell surface strongly suggest that the function(s) of this gene family may differ from those of other multigene families of Plasmodium, such as the var genes of Plasmodium falciparum.
Keywords: Plasmodium; acute infection; chronic infection; malaria; multigene families; pir genes.
Copyright © 2022 Giorgalli, Cunningham, Broncel, Sait, Harrison, Hosking, Vandomme, Amis, Antonello, Sullivan, Uwadiae, Torella, Higgins and Langhorne.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
-
- Atkinson C. T., Aikawa M. (1990). Ultrastructure of Malaria-Infected Erythrocytes. Blood Cells 16 (2-3), 351–368. - PubMed
-
- Baruch D. I., Gormely J. A., Ma C., Howard R. J., Pasloske B. L. (1996). Plasmodium Falciparum Erythrocyte Membrane Protein 1 is a Parasitised Erythrocyte Receptor for Adherence to CD36, Thrombospondin, and Intercellular Adhesion Molecule 1. Proc. Natl. Acad. Sci. U.S.A. 93 (8), 3497–3502. doi: 10.1073/pnas.93.8.3497 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

