Optimal Sacrificial Domains in Mechanical Polyproteins: S. epidermidis Adhesins Are Tuned for Work Dissipation
- PMID: 35783175
- PMCID: PMC9241160
- DOI: 10.1021/jacsau.2c00121
Optimal Sacrificial Domains in Mechanical Polyproteins: S. epidermidis Adhesins Are Tuned for Work Dissipation
Abstract
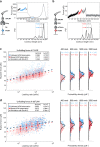
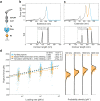
The opportunistic pathogen Staphylococcus epidermidis utilizes a multidomain surface adhesin protein to bind host components and adhere to tissues. While it is known that the interaction between the SdrG receptor and its fibrinopeptide target (FgB) is exceptionally mechanostable (∼2 nN), the influence of downstream B domains (B1 and B2) is unclear. Here, we studied the mechanical relationships between folded B domains and the SdrG receptor bound to FgB. We used protein engineering, single-molecule force spectroscopy (SMFS) with an atomic force microscope (AFM), and Monte Carlo simulations to understand how the mechanical properties of folded sacrificial domains, in general, can be optimally tuned to match the stability of a receptor-ligand complex. Analogous to macroscopic suspension systems, sacrificial shock absorber domains should neither be too weak nor too strong to optimally dissipate mechanical energy. We built artificial molecular shock absorber systems based on the nanobody (VHH) scaffold and studied the competition between domain unfolding and receptor unbinding. We quantitatively determined the optimal stability of shock absorbers that maximizes work dissipation on average for a given receptor and found that natural sacrificial domains from pathogenic S. epidermidis and Clostridium perfringens adhesins exhibit stabilities at or near this optimum within a specific range of loading rates. These findings demonstrate how tuning the stability of sacrificial domains in adhesive polyproteins can be used to maximize mechanical work dissipation and serve as an adhesion strategy by bacteria.
© 2022 The Authors. Published by American Chemical Society.
Conflict of interest statement
The authors declare no competing financial interest.
Figures



Similar articles
-
The binding force of the staphylococcal adhesin SdrG is remarkably strong.Mol Microbiol. 2014 Jul;93(2):356-68. doi: 10.1111/mmi.12663. Epub 2014 Jun 20. Mol Microbiol. 2014. PMID: 24898289
-
SdrG, a fibrinogen-binding bacterial adhesin of the microbial surface components recognizing adhesive matrix molecules subfamily from Staphylococcus epidermidis, targets the thrombin cleavage site in the Bbeta chain.J Biol Chem. 2001 Jul 27;276(30):27799-805. doi: 10.1074/jbc.M103873200. Epub 2001 May 22. J Biol Chem. 2001. PMID: 11371571
-
Staphylococcus epidermidis Affinity for Fibrinogen-Coated Surfaces Correlates with the Abundance of the SdrG Adhesin on the Cell Surface.Langmuir. 2015 Apr 28;31(16):4713-21. doi: 10.1021/acs.langmuir.5b00360. Epub 2015 Apr 13. Langmuir. 2015. PMID: 25821995
-
Single-molecule force spectroscopy on polyproteins and receptor-ligand complexes: The current toolbox.J Struct Biol. 2017 Jan;197(1):3-12. doi: 10.1016/j.jsb.2016.02.011. Epub 2016 Feb 9. J Struct Biol. 2017. PMID: 26873782 Review.
-
Next Generation Methods for Single-Molecule Force Spectroscopy on Polyproteins and Receptor-Ligand Complexes.Front Mol Biosci. 2020 May 19;7:85. doi: 10.3389/fmolb.2020.00085. eCollection 2020. Front Mol Biosci. 2020. PMID: 32509800 Free PMC article. Review.
Cited by
-
Engineering an artificial catch bond using mechanical anisotropy.Nat Commun. 2024 Apr 8;15(1):3019. doi: 10.1038/s41467-024-46858-9. Nat Commun. 2024. PMID: 38589360 Free PMC article.
-
S373P Mutation Stabilizes the Receptor-Binding Domain of the Spike Protein in Omicron and Promotes Binding.JACS Au. 2023 Jun 22;3(7):1902-1910. doi: 10.1021/jacsau.3c00142. eCollection 2023 Jul 24. JACS Au. 2023. PMID: 37502147 Free PMC article.
-
Unfolding of von Willebrand Factor Type D Like Domains Promotes Mucin Adhesion.Nano Lett. 2025 Feb 5;25(5):1765-1774. doi: 10.1021/acs.nanolett.4c03088. Epub 2025 Jan 22. Nano Lett. 2025. PMID: 39841791 Free PMC article.
-
Multi-state catch bond formed in the Izumo1:Juno complex that initiates human fertilization.Nat Commun. 2025 Aug 26;16(1):7952. doi: 10.1038/s41467-025-62427-0. Nat Commun. 2025. PMID: 40858532 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
