Characteristics, Biodiversity, and Cultivation Strategy of Low Nucleic Acid Content Bacteria
- PMID: 35783413
- PMCID: PMC9240426
- DOI: 10.3389/fmicb.2022.900669
Characteristics, Biodiversity, and Cultivation Strategy of Low Nucleic Acid Content Bacteria
Abstract
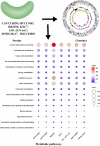
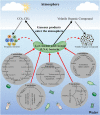
Low nucleic acid content (LNA) bacteria are ubiquitous and estimated to constitute 20%-90% of the total bacterial community in marine and freshwater environment. LNA bacteria with unique physiological characteristics, including small cell size and small genomes, can pass through 0.45-μm filtration. The researchers came up with different terminologies for low nucleic acid content bacteria based on different research backgrounds, such as: filterable bacteria, oligotrophic bacteria, and low-DNA bacteria. LNA bacteria have an extremely high level of genetic diversity and play an important role in material circulation in oligotrophic environment. However, the majority of LNA bacteria in the environment remain uncultivated. Thus, an important challenge now is to isolate more LNA bacteria from oligotrophic environments and gain insights into their unique metabolic mechanisms and ecological functions. Here, we reviewed LNA bacteria in aquatic environments, focusing on their characteristics, community structure and diversity, functions, and cultivation strategies. Exciting future prospects for LNA bacteria are also discussed.
Keywords: LNA bacteria; cultivation strategy; diversity; functions; physical characteristics; terminologies.
Copyright © 2022 Hu, Zhang, Lin, Liu, Bartlam and Wang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Metagenomics analysis reveals effects of salinity fluctuation on diversity and ecological functions of high and low nucleic acid content bacteria.Sci Total Environ. 2024 Jul 10;933:173186. doi: 10.1016/j.scitotenv.2024.173186. Epub 2024 May 12. Sci Total Environ. 2024. PMID: 38744390
-
Impact of planktonic low nucleic acid-content bacteria to bacterial community structure and associated ecological functions in a shallow lake.Sci Total Environ. 2019 Mar 25;658:868-878. doi: 10.1016/j.scitotenv.2018.12.274. Epub 2018 Dec 19. Sci Total Environ. 2019. PMID: 30678021
-
Isolation and characterization of low nucleic acid (LNA)-content bacteria.ISME J. 2009 Aug;3(8):889-902. doi: 10.1038/ismej.2009.46. Epub 2009 May 7. ISME J. 2009. PMID: 19421234
-
Size Matters: Ultra-small and Filterable Microorganisms in the Environment.Microbes Environ. 2020;35(2):ME20025. doi: 10.1264/jsme2.ME20025. Microbes Environ. 2020. PMID: 32493880 Free PMC article. Review.
-
[Isolation and characterization of marine oligotrophic bacteria].J Soc Biol. 2007;201(1):41-50. doi: 10.1051/jbio:2007005. J Soc Biol. 2007. PMID: 17762823 Review. French.
Cited by
-
Shaping the future of tobacco through microbial insights: a review of advances and applications.Front Bioeng Biotechnol. 2025 May 12;13:1548323. doi: 10.3389/fbioe.2025.1548323. eCollection 2025. Front Bioeng Biotechnol. 2025. PMID: 40421115 Free PMC article. Review.
-
Effects of Salinity Fluctuation on Antimicrobial Resistance and Virulence Factor Genes of Low and High Nucleic Acid-Content Bacteria in a Marine Environment.Microorganisms. 2025 Jul 21;13(7):1710. doi: 10.3390/microorganisms13071710. Microorganisms. 2025. PMID: 40732219 Free PMC article.
-
Spatiotemporal dynamics of high and low nucleic acid-content bacterial communities in Chinese coastal seawater: assembly process, co-occurrence relationship and the ecological functions.Front Microbiol. 2023 Aug 2;14:1219655. doi: 10.3389/fmicb.2023.1219655. eCollection 2023. Front Microbiol. 2023. PMID: 37601370 Free PMC article.
References
-
- Andrade L., Gonzalez A. M., Rezende C. E., Suzuki M., Valentin J. L., Paranhos R. (2007). Distribution of HNA and LNA bacterial groups in the Southwest Atlantic Ocean. Braz. J. Microbiol. 38, 330–336. doi: 10.1590/S1517-83822007000200028 - DOI
-
- Belzile C., Brugel S., Nozais C., Gratton Y., Demers S. (2008). Variations of the abundance and nucleic acid content of heterotrophic bacteria in Beaufort shelf waters during winter and spring. J. Mar. Syst. 74, 946–956. doi: 10.1016/j.jmarsys.2007.12.010 - DOI
-
- Cavicchioli R., Ostrowski M., Fegatella F., Goodchild A., Guixa-Boixereu N. (2003). Life under nutrient limitation in oligotrophic marine environments: an eco/physiological perspective of Sphingopyxis alaskensis (formerly Sphingomonas alaskensis). Microb. Ecol. 45, 203–217. doi: 10.1007/s00248-002-3008-6 - DOI - PubMed
Publication types
LinkOut - more resources
Full Text Sources

