Distinct Patterns of Rhizosphere Microbiota Associated With Rice Genotypes Differing in Aluminum Tolerance in an Acid Sulfate Soil
- PMID: 35783428
- PMCID: PMC9247542
- DOI: 10.3389/fmicb.2022.933722
Distinct Patterns of Rhizosphere Microbiota Associated With Rice Genotypes Differing in Aluminum Tolerance in an Acid Sulfate Soil
Abstract
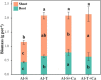
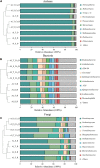
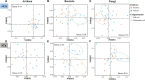
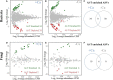
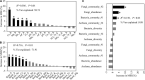
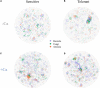
Rhizosphere microbes are important for plant tolerance to various soil stresses. Rice is the most aluminum (Al)-tolerant small grain cereal crop species, but the link between rice Al tolerance and rhizosphere microbiota remains unclear. This study aimed to investigate the microbial community structure of aluminum-sensitive and Al-tolerant rice varieties in acid sulfate soil under liming and non-liming conditions. We analyzed the rice biomass and mineral element contents of rice plants as well as the chemical properties and microbial (archaea, bacteria, and fungi) communities of rhizosphere and bulk soil samples. The results showed that the Al-tolerant rice genotype grew better and was able to take up more phosphorus from the acid sulfate soil than the Al-sensitive genotype. Liming was the main factor altering the microbial diversity and community structure, followed by rhizosphere effects. In the absence of liming effects, the rice genotypes shifted the community structure of bacteria and fungi, which accounted for the observed variation in the rice biomass. The Al-tolerant rice genotype recruited specific bacterial and fungal taxa (Bacillus, Pseudomonas, Aspergillus, and Rhizopus) associated with phosphorus solubilization and plant growth promotion. The soil microbial co-occurrence network of the Al-tolerant rice genotype was more complex than that of the Al-sensitive rice genotype. In conclusion, the bacterial and fungal community in the rhizosphere has genotype-dependent effects on rice Al tolerance. Aluminum-tolerant rice genotypes recruit specific microbial taxa, especially phosphorus-solubilizing microorganisms, and are associated with complex microbial co-occurrence networks, which may enhance rice growth in acid sulfate soil.
Keywords: acid sulfate soil; aluminum toxicity; co-occurrence network; microbial community structure; rice genotypes.
Copyright © 2022 Xiao, Wang, Li, Li, Dai, Shen and Zhao.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Assemblage of root-associated microbiome contributes to disparate performance of two rice genotypes under aluminum stress.Plant Physiol Biochem. 2025 Mar;220:109539. doi: 10.1016/j.plaphy.2025.109539. Epub 2025 Jan 29. Plant Physiol Biochem. 2025. PMID: 39908935
-
Rhizosphere Soil Fungal Communities of Aluminum-Tolerant and -Sensitive Soybean Genotypes Respond Differently to Aluminum Stress in an Acid Soil.Front Microbiol. 2020 May 28;11:1177. doi: 10.3389/fmicb.2020.01177. eCollection 2020. Front Microbiol. 2020. PMID: 32547532 Free PMC article.
-
Recruitment of specific rhizosphere microorganisms in saline-alkali tolerant rice improves adaptation to saline-alkali stress.Sci Total Environ. 2025 Feb 1;963:178413. doi: 10.1016/j.scitotenv.2025.178413. Epub 2025 Jan 17. Sci Total Environ. 2025. PMID: 39824093
-
Bacterial and fungal communities in bulk soil and rhizospheres of aluminum-tolerant and aluminum-sensitive maize (Zea mays L.) lines cultivated in unlimed and limed Cerrado soil.J Microbiol Biotechnol. 2008 May;18(5):805-14. J Microbiol Biotechnol. 2008. PMID: 18633275
-
Aluminum in plant: Benefits, toxicity and tolerance mechanisms.Front Plant Sci. 2023 Jan 13;13:1085998. doi: 10.3389/fpls.2022.1085998. eCollection 2022. Front Plant Sci. 2023. PMID: 36714730 Free PMC article. Review.
Cited by
-
Soil bacterial communities of paddy are dependent on root compartment niches but independent of growth stages from Mollisols of Northeast China.Front Microbiol. 2023 Apr 14;14:1170611. doi: 10.3389/fmicb.2023.1170611. eCollection 2023. Front Microbiol. 2023. PMID: 37125155 Free PMC article.
-
The Rice-Microbe Nexus: Unlocking Productivity Through Soil Science.Rice (N Y). 2025 Jun 20;18(1):56. doi: 10.1186/s12284-025-00809-0. Rice (N Y). 2025. PMID: 40540085 Free PMC article. Review.
-
Salt stress alters the selectivity of mature pecan for the rhizosphere community and its associated functional traits.Front Plant Sci. 2025 Mar 26;16:1473473. doi: 10.3389/fpls.2025.1473473. eCollection 2025. Front Plant Sci. 2025. PMID: 40206877 Free PMC article.
-
Continuous-cropping-tolerant soybean cultivars alleviate continuous cropping obstacles by improving structure and function of rhizosphere microorganisms.Front Microbiol. 2023 Jan 4;13:1048747. doi: 10.3389/fmicb.2022.1048747. eCollection 2022. Front Microbiol. 2023. PMID: 36687563 Free PMC article.
-
Stochastic processes drive the soil fungal communities in a developing mid-channel bar.Front Microbiol. 2023 Feb 6;14:1104297. doi: 10.3389/fmicb.2023.1104297. eCollection 2023. Front Microbiol. 2023. PMID: 36814566 Free PMC article.
References
-
- Andrews S. (2010). FastQC: a quality control tool for high throughput sequence data. Version 0.11.2. Website Httpwww Bioin Forma Tics Babra Ham Ac Ukproje Ctsfastqc.
-
- Ayangbenro A. S., Babalola O. O. (2021). Reclamation of arid and semi-arid soils: The role of plant growth-promoting archaea and bacteria. Curr. Plant Biol. 25 100173. 10.1016/j.cpb.2020.100173 - DOI
-
- Bastian M., Heymann S., Jacomy M. (2009). “Gephi: an open source software for exploring and manipulating networks,” in International AAAI conference on weblogs and social media. (Association for the Advancement of Artificial Intelligence; ). 2009 361–362.
LinkOut - more resources
Full Text Sources
Other Literature Sources

