The Chromosome-Scale Assembly of the Curcuma alismatifolia Genome Provides Insight Into Anthocyanin and Terpenoid Biosynthesis
- PMID: 35783929
- PMCID: PMC9241516
- DOI: 10.3389/fpls.2022.899588
The Chromosome-Scale Assembly of the Curcuma alismatifolia Genome Provides Insight Into Anthocyanin and Terpenoid Biosynthesis
Abstract
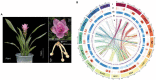
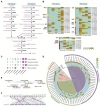
Curcuma alismatifolia, a bulbous flower known for its showy bracts, is widely used around the world as a cut flower, potted, and garden plant. Besides its ornamental value, this species is rich in terpenoid metabolites and could serve as a resource for essential oils. Here, we report a chromosome-level genome assembly of C. alismatifolia and describe its biosynthetic pathways for anthocyanins and terpenoids. This high-quality, assembled genome size is 991.3 Mb with a scaffold N50 value of 56.7 Mb. Evolutionary analysis of the genome suggests that C. alismatifolia diverged from Zingiber officinale about 9.7 million years ago, after it underwent a whole-genome duplication. Transcriptome analysis was performed on bracts at five developmental stages. Nine highly expressed genes were identified, encoding for six enzymes downstream of the anthocyanin biosynthetic pathway. Of these, one gene encoding F3'5'H might be a key node in the regulation of bract color formation. Co-expression network analysis showed that MYB, bHLH, NAC, and ERF transcription factors collectively regulated color formation in the bracts. Characterization of terpenoid biosynthesis genes revealed their dispersal and tandem duplications, both of which contributed greatly to the increase in the number of terpene synthase genes in C. alismatifolia, especially to species-specific expansion of sesquiterpene synthase genes. This work facilitates understanding of genetic basis of anthocyanin and terpenoid biosynthesis and could accelerate the selective breeding of C. alismatifolia varieties with higher ornamental and medicinal value.
Keywords: C. alismatifolia; anthocyanin; evolution; genome; terpenoid.
Copyright © 2022 Dong, Zou, Mao, Tian, Hu, Cao and Ding.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Role of anthocyanin metabolic diversity in bract coloration of Curcuma alismatifolia varieties.Plant Physiol Biochem. 2024 Nov;216:109156. doi: 10.1016/j.plaphy.2024.109156. Epub 2024 Sep 26. Plant Physiol Biochem. 2024. PMID: 39341180
-
The genomic and bulked segregant analysis of Curcuma alismatifolia revealed its diverse bract pigmentation.aBIOTECH. 2022 Oct 6;3(3):178-196. doi: 10.1007/s42994-022-00081-6. eCollection 2022 Sep. aBIOTECH. 2022. PMID: 36304840 Free PMC article.
-
Transcriptome Analysis Revealed the Anabolic Regulation of Chlorophyll and Carotenoids in Curcuma alismatifolia Bracts.Biochem Genet. 2024 Sep 26. doi: 10.1007/s10528-024-10923-1. Online ahead of print. Biochem Genet. 2024. PMID: 39327377
-
Flower colour and cytochromes P450.Philos Trans R Soc Lond B Biol Sci. 2013 Jan 6;368(1612):20120432. doi: 10.1098/rstb.2012.0432. Print 2013 Feb 19. Philos Trans R Soc Lond B Biol Sci. 2013. PMID: 23297355 Free PMC article. Review.
-
Advances in the Biosynthesis of Plant Terpenoids: Models, Mechanisms, and Applications.Plants (Basel). 2025 May 10;14(10):1428. doi: 10.3390/plants14101428. Plants (Basel). 2025. PMID: 40430993 Free PMC article. Review.
Cited by
-
Combined Metabolome and Transcriptome Analysis of Creamy Yellow and Purple Colored Panax notoginseng Roots.Life (Basel). 2023 Oct 23;13(10):2100. doi: 10.3390/life13102100. Life (Basel). 2023. PMID: 37895482 Free PMC article.
-
Metabolomic and transcriptomic analyses provide insight into the variation of floral scent and molecular regulation in different cultivars and flower development of Curcuma alismatifolia.Hortic Res. 2024 Dec 12;12(3):uhae348. doi: 10.1093/hr/uhae348. eCollection 2025 Mar. Hortic Res. 2024. PMID: 40061800 Free PMC article.
-
Advances in Bract Coloration: Diversity, Pigment Synthesis, and Regulatory Mechanisms in Ornamental Plants.Plants (Basel). 2025 Jul 13;14(14):2155. doi: 10.3390/plants14142155. Plants (Basel). 2025. PMID: 40733391 Free PMC article. Review.
-
Multiomics comparison among populations of three plant sources of Amomi Fructus.Hortic Res. 2023 Aug 1;10(8):uhad128. doi: 10.1093/hr/uhad128. eCollection 2023 Aug. Hortic Res. 2023. PMID: 37560015 Free PMC article.
-
Small cardamom genome: development and utilization of microsatellite markers from a draft genome sequence of Elettaria cardamomum Maton.Front Plant Sci. 2023 May 10;14:1161499. doi: 10.3389/fpls.2023.1161499. eCollection 2023. Front Plant Sci. 2023. PMID: 37235027 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Miscellaneous

