Sphingolipid Long-Chain Base Phosphate Degradation Can Be a Rate-Limiting Step in Long-Chain Base Homeostasis
- PMID: 35783987
- PMCID: PMC9240600
- DOI: 10.3389/fpls.2022.911073
Sphingolipid Long-Chain Base Phosphate Degradation Can Be a Rate-Limiting Step in Long-Chain Base Homeostasis
Abstract
Sphingolipid long-chain bases (LCBs) are building blocks for membrane-localized sphingolipids, and are involved in signal transduction pathways in plants. Elevated LCB levels are associated with the induction of programmed cell death and pathogen-derived toxin-induced cell death. Therefore, levels of free LCBs can determine survival of plant cells. To elucidate the contribution of metabolic pathways regulating high LCB levels, we applied the deuterium-labeled LCB D-erythro-sphinganine-d7 (D7-d18:0), the first LCB in sphingolipid biosynthesis, to Arabidopsis leaves and quantified labeled LCBs, LCB phosphates (LCB-Ps), and 14 abundant ceramide (Cer) species over time. We show that LCB D7-d18:0 is rapidly converted into the LCBs d18:0P, t18:0, and t18:0P. Deuterium-labeled ceramides were less abundant, but increased over time, with the highest levels detected for Cer(d18:0/16:0), Cer(d18:0/24:0), Cer(t18:0/16:0), and Cer(t18:0/22:0). A more than 50-fold increase of LCB-P levels after leaf incubation in LCB D7-d18:0 indicated that degradation of LCBs via LCB-Ps is important, and we hypothesized that LCB-P degradation could be a rate-limiting step to reduce high levels of LCBs. To functionally test this hypothesis, we constructed a transgenic line with dihydrosphingosine-1-phosphate lyase 1 (DPL1) under control of an inducible promotor. Higher expression of DPL1 significantly reduced elevated LCB-P and LCB levels induced by Fumonisin B1, and rendered plants more resistant against this fungal toxin. Taken together, we provide quantitative data on the contribution of major enzymatic pathways to reduce high LCB levels, which can trigger cell death. Specifically, we provide functional evidence that DPL1 can be a rate-limiting step in regulating high LCB levels.
Keywords: LC–MS/MS; cell death; dihydrosphingosine-1-phosphate lyase; long-chain base; metabolic flux analysis; plant sphingolipid metabolism; sphingolipid.
Copyright © 2022 Lambour, Glenz, Forner, Krischke, Mueller, Fekete and Waller.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

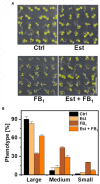


References
-
- Alden K. P., Dhondt-Cordelier S., McDonald K. L., Reape T. J., Ng C. K. Y., McCabe P. F., et al. (2011). Sphingolipid long chain base phosphates can regulate apoptotic-like programmed cell death in plants. Biochem. Biophys. Res. Commun. 410, 574–580. doi: 10.1016/j.bbrc.2011.06.028, PMID: - DOI - PubMed
-
- Chen M., Cahoon E., Saucedo-García M., Plasencia J., Gavilanes-Ruiz M. (2009). “Plant Sphingolipids: Structure, Synthesis and Function,” in Lipids in Photosynthesis. Advances in Photosynthesis and Respiration. ed. Wada M. N. H. (Dordrecht: Springer; ), 77–115.
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

