Gut Mycobiome in Patients With Chronic Kidney Disease Was Altered and Associated With Immunological Profiles
- PMID: 35784313
- PMCID: PMC9245424
- DOI: 10.3389/fimmu.2022.843695
Gut Mycobiome in Patients With Chronic Kidney Disease Was Altered and Associated With Immunological Profiles
Abstract
Objectives: Mounting evidence suggests that bacterial dysbiosis and immunity disorder are associated with patients with chronic kidney disease (CKD), but the mycobiome is beginning to gain recognition as a fundamental part of our microbiome. We aim to characterize the profile of the mycobiome in the gut of CKD patients and its correlation to serum immunological profiles.
Methods and materials: Ninety-two CKD patients and sex-age-body mass index (BMI)-matched healthy controls (HCs) were recruited. Fresh samples were collected using sterile containers. ITS transcribed spacer ribosomal RNA gene sequencing was performed on the samples. An immunoturbidimetric test was used to assess the serum levels of immunological features.
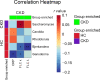
Results: The CKD cohort displayed a different microbial community from that in the HC cohort according to principal coordinate analysis (PCoA). (P=0.001). The comparison of the two cohorts showed that the CKD cohort had significantly higher gut microbial richness and diversity (P<0.05). The CKD cohort had lower abundances of Candida, Bjerkandera, Rhodotorula, and Ganoderma compared to the HC cohort, while it had higher Saccharomyces (P<0.05). However, the microbial community alteration was inconsistent with the severity of kidney damage in patients, as only patients in CKD stage 1~3 had differed microbial community concerning for HCs based on PCoA (P<0.05). The serum concentration of the kappa light chain in CKD patients was positively associated with Saccharomyces, whereas the it was negatively associated with Ganoderma (P<0.05).
Conclusions: Not only was gut mycobiome dysbiosis observed in CKD patients, but the dysbiosis was also associated with the immunological disorder. These findings suggest that therapeutic strategies targeting gut mycobiome might be effective.
Keywords: Candida; Saccharomyces; chronic kidney disease; immunity disorder; microbial dysbiosis; mycobiome.
Copyright © 2022 Hu, Wei, Gu, Wang, Feng, Sheng, Hu, Gu, Jiang, Tian, Guo, Lv, Liu, Zou, Yan and Feng.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Exploring the gut mycobiome: differential composition and clinical associations in hypertension, chronic kidney disease, and their comorbidity.Front Immunol. 2023 Dec 14;14:1317809. doi: 10.3389/fimmu.2023.1317809. eCollection 2023. Front Immunol. 2023. PMID: 38162661 Free PMC article.
-
Gut mycobiome dysbiosis contributes to the development of hypertension and its response to immunoglobulin light chains.Front Immunol. 2022 Dec 29;13:1089295. doi: 10.3389/fimmu.2022.1089295. eCollection 2022. Front Immunol. 2022. PMID: 36643913 Free PMC article.
-
Alteration of the Gut Microbiome in Chronic Kidney Disease Patients and Its Association With Serum Free Immunoglobulin Light Chains.Front Immunol. 2021 Apr 1;12:609700. doi: 10.3389/fimmu.2021.609700. eCollection 2021. Front Immunol. 2021. PMID: 33868230 Free PMC article.
-
Specific alterations in gut microbiota in patients with chronic kidney disease: an updated systematic review.Ren Fail. 2021 Dec;43(1):102-112. doi: 10.1080/0886022X.2020.1864404. Ren Fail. 2021. PMID: 33406960 Free PMC article.
-
The clinical impact of gut microbiota in chronic kidney disease.Korean J Intern Med. 2020 Nov;35(6):1305-1316. doi: 10.3904/kjim.2020.411. Epub 2020 Sep 29. Korean J Intern Med. 2020. PMID: 32872729 Free PMC article. Review.
Cited by
-
Exploring the gut mycobiome: differential composition and clinical associations in hypertension, chronic kidney disease, and their comorbidity.Front Immunol. 2023 Dec 14;14:1317809. doi: 10.3389/fimmu.2023.1317809. eCollection 2023. Front Immunol. 2023. PMID: 38162661 Free PMC article.
-
The evolving understanding of systemic mechanisms in organ-specific IgA nephropathy: a focus on gut-kidney crosstalk.Theranostics. 2025 Jan 1;15(2):656-681. doi: 10.7150/thno.104631. eCollection 2025. Theranostics. 2025. PMID: 39744688 Free PMC article. Review.
-
The compositional and functional imbalance of the gut microbiota in CKD linked to disease patterns.J Transl Med. 2024 Aug 16;22(1):773. doi: 10.1186/s12967-024-05578-w. J Transl Med. 2024. PMID: 39152439 Free PMC article.
-
Causal relationship between gut microbiota and chronic renal failure: a two-sample Mendelian randomization study.Front Microbiol. 2024 Apr 3;15:1356478. doi: 10.3389/fmicb.2024.1356478. eCollection 2024. Front Microbiol. 2024. PMID: 38633704 Free PMC article.
-
The Gut Mycobiome for Precision Medicine.J Fungi (Basel). 2025 Apr 2;11(4):279. doi: 10.3390/jof11040279. J Fungi (Basel). 2025. PMID: 40278100 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

