Importance of Rare DPYD Genetic Polymorphisms for 5-Fluorouracil Therapy in the Japanese Population
- PMID: 35784703
- PMCID: PMC9242541
- DOI: 10.3389/fphar.2022.930470
Importance of Rare DPYD Genetic Polymorphisms for 5-Fluorouracil Therapy in the Japanese Population
Abstract
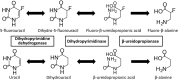
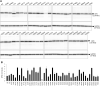
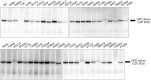
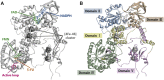
Dihydropyrimidine dehydrogenase (DPD), encoded by the DPYD gene, is the rate-limiting enzyme in 5-fluorouracil (5-FU) degradation. In Caucasians, four DPYD risk variants are recognized to be responsible for interindividual variations in the development of 5-FU toxicity. However, these risk variants have not been identified in Asian populations. Recently, 41 DPYD allelic variants, including 15 novel single nucleotide variants, were identified in 3,554 Japanese individuals by analyzing their whole-genome sequences; however, the effects of these variants on DPD enzymatic activity remain unknown. In the present study, an in vitro analysis was performed on 41 DPD allelic variants and three DPD risk variants to elucidate the changes in enzymatic activity. Wild-type and 44 DPD-variant proteins were heterologously expressed in 293FT cells. DPD expression levels and dimerization of DPD were determined by immunoblotting after SDS-PAGE and blue native PAGE, respectively. The enzymatic activity of DPD was evaluated by quantification of dihydro-5-FU, a metabolite of 5-FU, using high-performance liquid chromatography-tandem mass spectrometry. Moreover, we used 3D simulation modeling to analyze the effect of amino acid substitutions on the conformation of DPD. Among the 41 DPD variants, seven exhibited drastically decreased intrinsic clearance (CL int ) compared to the wild-type protein. Moreover, R353C and G926V exhibited no enzymatic activity, and the band patterns observed in the immunoblots after blue native PAGE indicated that DPD dimerization is required for its enzymatic activity. Our data suggest that these variants may contribute to the significant inter-individual variability observed in the pharmacokinetics and pharmacodynamics of 5-FU. In our study, nine DPD variants exhibited drastically decreased or no enzymatic activity due to dimerization inhibition or conformational changes in each domain. Especially, the rare DPYD variants, although at very low frequencies, may serve as important pharmacogenomic markers associated with the severe 5-FU toxicity in Japanese population.
Keywords: 5-fluorouracil; DPYD; dihydropyrimidine dehydrogenase (DPD); genetic polymorphism; pharmacogenomics.
Copyright © 2022 Hishinuma, Narita, Obuchi, Ueda, Saito, Tadaka, Kinoshita, Maekawa, Mano, Hirasawa and Hiratsuka.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Functional Characterization of 21 Allelic Variants of Dihydropyrimidine Dehydrogenase Identified in 1070 Japanese Individuals.Drug Metab Dispos. 2018 Aug;46(8):1083-1090. doi: 10.1124/dmd.118.081737. Epub 2018 May 16. Drug Metab Dispos. 2018. PMID: 29769267
-
Comparative functional analysis of DPYD variants of potential clinical relevance to dihydropyrimidine dehydrogenase activity.Cancer Res. 2014 May 1;74(9):2545-54. doi: 10.1158/0008-5472.CAN-13-2482. Epub 2014 Mar 19. Cancer Res. 2014. PMID: 24648345 Free PMC article.
-
Predicting 5-fluorouracil toxicity: DPD genotype and 5,6-dihydrouracil:uracil ratio.Pharmacogenomics. 2014 Sep;15(13):1653-66. doi: 10.2217/pgs.14.126. Pharmacogenomics. 2014. PMID: 25410891
-
Clinical implications of dihydropyrimidine dehydrogenase (DPD) activity in 5-FU-based chemotherapy: mutations in the DPD gene, and DPD inhibitory fluoropyrimidines.Int J Clin Oncol. 2003 Jun;8(3):132-8. doi: 10.1007/s10147-003-0330-z. Int J Clin Oncol. 2003. PMID: 12851836 Review.
-
Implications of dihydropyrimidine dehydrogenase on 5-fluorouracil pharmacogenetics and pharmacogenomics.Pharmacogenomics. 2002 Jul;3(4):485-92. doi: 10.1517/14622416.3.4.485. Pharmacogenomics. 2002. PMID: 12164772 Review.
Cited by
-
Poor association between dihydropyrimidine dehydrogenase (DPYD) genotype and fluoropyrimidine-induced toxicity in an Asian population.Cancer Med. 2023 Apr;12(7):7808-7814. doi: 10.1002/cam4.5541. Epub 2022 Dec 16. Cancer Med. 2023. PMID: 36524458 Free PMC article.
-
Frequencies of pharmacogenomic alleles across biogeographic groups in a large-scale biobank.Am J Hum Genet. 2023 Oct 5;110(10):1628-1647. doi: 10.1016/j.ajhg.2023.09.001. Epub 2023 Sep 26. Am J Hum Genet. 2023. PMID: 37757824 Free PMC article.
-
Outcomes of capecitabine plus temozolomide combination therapy in patients with advanced or metastatic pancreatic neuroendocrine tumors: a retrospective observational single-center study.Int J Clin Oncol. 2025 Jul;30(7):1409-1416. doi: 10.1007/s10147-025-02779-1. Epub 2025 May 9. Int J Clin Oncol. 2025. PMID: 40346312
-
DPYD genetic polymorphisms in non-European patients with severe fluoropyrimidine-related toxicity: a systematic review.Br J Cancer. 2024 Aug;131(3):498-514. doi: 10.1038/s41416-024-02754-z. Epub 2024 Jun 17. Br J Cancer. 2024. PMID: 38886557 Free PMC article.
-
A Systematic Review and Meta-Analysis of Trifluridine/Tipiracil plus Bevacizumab for the Treatment of Metastatic Colorectal Cancer: Evidence from Real-World Series.Curr Oncol. 2023 May 24;30(6):5227-5239. doi: 10.3390/curroncol30060397. Curr Oncol. 2023. PMID: 37366880 Free PMC article.
References
-
- Amstutz U., Henricks L. M., Offer S. M., Barbarino J., Schellens J. H. M., Swen J. J., et al. (2018). Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for Dihydropyrimidine Dehydrogenase Genotype and Fluoropyrimidine Dosing: 2017 Update. Clin. Pharmacol. Ther. 103 (2), 210–216. 10.1002/cpt.911 - DOI - PMC - PubMed
-
- Brečević L., Rinčić M., Krsnik Ž., Sedmak G., Hamid A. B., Kosyakova N., et al. (2015). Association of New Deletion/duplication Region at Chromosome 1p21 with Intellectual Disability, Severe Speech Deficit and Autism Spectrum Disorder-like Behavior: an All-In Approach to Solving the DPYD Enigma. Transl. Neurosci. 6 (1), 59–86. 10.1515/tnsci-2015-0007 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

