DNA Strand-Displacement Temporal Logic Circuits
- PMID: 35785961
- PMCID: PMC9284558
- DOI: 10.1021/jacs.2c04325
DNA Strand-Displacement Temporal Logic Circuits
Abstract
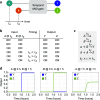
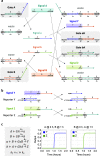
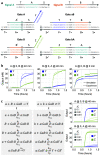
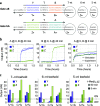
Molecular circuits capable of processing temporal information are essential for complex decision making in response to both the presence and history of a molecular environment. A particular type of temporal information that has been recognized to be important is the relative timing of signals. Here we demonstrate the strategy of temporal memory combined with logic computation in DNA strand-displacement circuits capable of making decisions based on specific combinations of inputs as well as their relative timing. The circuit encodes the timing information on inputs in a set of memory strands, which allows for the construction of logic gates that act on current and historical signals. We show that mismatches can be employed to reduce the complexity of circuit design and that shortening specific toeholds can be useful for improving the robustness of circuit behavior. We also show that a detailed model can provide critical insights for guiding certain aspects of experimental investigations that an abstract model cannot. We envision that the design principles explored in this study can be generalized to more complex temporal logic circuits and incorporated into other types of circuit architectures, including DNA-based neural networks, enabling the implementation of timing-dependent learning rules and opening up new opportunities for embedding intelligent behaviors into artificial molecular machines.
Conflict of interest statement
The authors declare no competing financial interest.
Figures

Similar articles
-
A molecular assessment of the practical potential of DNA-based computation.Curr Opin Biotechnol. 2023 Jun;81:102940. doi: 10.1016/j.copbio.2023.102940. Epub 2023 Apr 13. Curr Opin Biotechnol. 2023. PMID: 37058876 Free PMC article. Review.
-
Fast and compact DNA logic circuits based on single-stranded gates using strand-displacing polymerase.Nat Nanotechnol. 2019 Nov;14(11):1075-1081. doi: 10.1038/s41565-019-0544-5. Epub 2019 Sep 23. Nat Nanotechnol. 2019. PMID: 31548688
-
Scaling Up Multi-bit DNA Full Adder Circuits with Minimal Strand Displacement Reactions.J Am Chem Soc. 2022 Jun 1;144(21):9479-9488. doi: 10.1021/jacs.2c03258. Epub 2022 May 22. J Am Chem Soc. 2022. PMID: 35603742
-
Mapping Temporally Ordered Inputs to Binary Message Outputs with a DNA Temporal Logic Circuit.Nanomaterials (Basel). 2023 Feb 27;13(5):903. doi: 10.3390/nano13050903. Nanomaterials (Basel). 2023. PMID: 36903782 Free PMC article.
-
Boolean logic gate based on DNA strand displacement for biosensing: current and emerging strategies.Nanoscale Horiz. 2021 Apr 1;6(4):298-310. doi: 10.1039/d0nh00587h. Epub 2021 Mar 24. Nanoscale Horiz. 2021. PMID: 33877218 Review.
Cited by
-
A molecular assessment of the practical potential of DNA-based computation.Curr Opin Biotechnol. 2023 Jun;81:102940. doi: 10.1016/j.copbio.2023.102940. Epub 2023 Apr 13. Curr Opin Biotechnol. 2023. PMID: 37058876 Free PMC article. Review.
-
High-Speed Sequential DNA Computing Using a Solid-State DNA Origami Register.ACS Cent Sci. 2024 Dec 11;10(12):2285-2293. doi: 10.1021/acscentsci.4c01557. eCollection 2024 Dec 25. ACS Cent Sci. 2024. PMID: 39735316 Free PMC article.
-
DNA-based customized functional modules for signal transformation.Front Chem. 2023 Feb 14;11:1140022. doi: 10.3389/fchem.2023.1140022. eCollection 2023. Front Chem. 2023. PMID: 36864900 Free PMC article. Review.
-
An Exo III-powered closed-loop DNA circuit architecture for biosensing/imaging.Mikrochim Acta. 2024 Jun 14;191(7):395. doi: 10.1007/s00604-024-06476-0. Mikrochim Acta. 2024. PMID: 38877347
-
A Localized Scalable DNA Logic Circuit System Based on the DNA Origami Surface.Int J Mol Sci. 2025 Feb 26;26(5):2043. doi: 10.3390/ijms26052043. Int J Mol Sci. 2025. PMID: 40076663 Free PMC article.
References
-
- Lakin M. R.; Stefanovic D.. Towards temporal logic computation using DNA strand displacement reactions. Unconventional Computation and Natural Computation; 16th International Conference, UCNC 2017, Fayetteville, AR, USA, June 5–9, 2017, Springer, 2017; pp 41–55.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

