Emerging Themes in CryoEM─Single Particle Analysis Image Processing
- PMID: 35785962
- PMCID: PMC9479088
- DOI: 10.1021/acs.chemrev.1c00850
Emerging Themes in CryoEM─Single Particle Analysis Image Processing
Abstract
Cryo-electron microscopy (CryoEM) has become a vital technique in structural biology. It is an interdisciplinary field that takes advantage of advances in biochemistry, physics, and image processing, among other disciplines. Innovations in these three basic pillars have contributed to the boosting of CryoEM in the past decade. This work reviews the main contributions in image processing to the current reconstruction workflow of single particle analysis (SPA) by CryoEM. Our review emphasizes the time evolution of the algorithms across the different steps of the workflow differentiating between two groups of approaches: analytical methods and deep learning algorithms. We present an analysis of the current state of the art. Finally, we discuss the emerging problems and challenges still to be addressed in the evolution of CryoEM image processing methods in SPA.
Conflict of interest statement
The authors declare no competing financial interest.
Figures

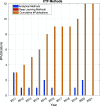
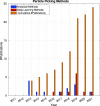
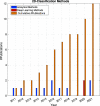
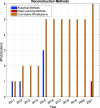
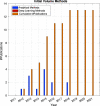
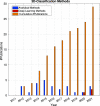
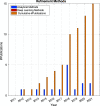
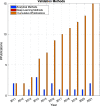
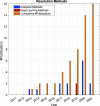
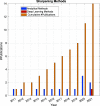
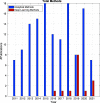
References
-
- Kai J.Zhang – Gautomatch. https://www2.mrc-lmb.cam.ac.uk/research/locally-developed-software/zhang... (accessed 2018-05-16).
-
- Abrishami V.; Zaldívar-Peraza A.; de la Rosa-Trevín J. M.; Vargas J.; Otón J.; Marabini R.; Shkolnisky Y.; Carazo J. M.; Sorzano C. O. S. A pattern matching approach to the automatic selection of particles from low-contrast electron micrographs. Bioinformatics 2013, 29, 2460–2468. 10.1093/bioinformatics/btt429. - DOI - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

