Genomic map of candidate human imprint control regions: the imprintome
- PMID: 35786392
- PMCID: PMC9665137
- DOI: 10.1080/15592294.2022.2091815
Genomic map of candidate human imprint control regions: the imprintome
Abstract
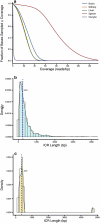
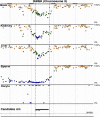
Imprinted genes - critical for growth, metabolism, and neuronal function - are expressed from one parental allele. Parent-of-origin-dependent CpG methylation regulates this expression at imprint control regions (ICRs). Since ICRs are established before tissue specification, these methylation marks are similar across cell types. Thus, they are attractive for investigating the developmental origins of adult diseases using accessible tissues, but remain unknown. We determined genome-wide candidate ICRs in humans by performing whole-genome bisulphite sequencing (WGBS) of DNA derived from the three germ layers and from gametes. We identified 1,488 hemi-methylated candidate ICRs, including 19 of 25 previously characterized ICRs (https://humanicr.org/). Gamete methylation approached 0% or 100% in 332 ICRs (178 paternally and 154 maternally methylated), supporting parent-of-origin-specific methylation, and 65% were in well-described CTCF-binding or DNaseI hypersensitive regions. This draft of the human imprintome will allow for the systematic determination of the role of early-acquired imprinting dysregulation in the pathogenesis of human diseases and developmental and behavioural disorders.
Keywords: Epigenetics; foetal origins; genomic imprinting; imprint control regions; methylation; whole genome.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures


References
-
- Badcock C, Crespi B.. Battle of the sexes may set the brain. Nature. 2008;454(7208):1054–1055. - PubMed
-
- Jirtle RL. Genomic imprinting and cancer. Exp Cell Res. 1999;248(1):18–24. - PubMed
-
- Jirtle RL. IGF2 loss of imprinting: a potential heritable risk factor for colorectal cancer. Gastroenterology. 2004;126(4):1190–1193. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
