TF-DUBTACs Stabilize Tumor Suppressor Transcription Factors
- PMID: 35786952
- PMCID: PMC10981454
- DOI: 10.1021/jacs.2c04824
TF-DUBTACs Stabilize Tumor Suppressor Transcription Factors
Abstract
Targeted protein degradation approaches have been widely used for degrading oncogenic proteins, providing a potentially promising therapeutic strategy for cancer treatment. However, approaches to targeting tumor suppressor proteins are very limited, and only a few agonists have been developed to date. Here, we report the development of a platform termed TF-DUBTAC, which links a DNA oligonucleotide to a covalent ligand of the deubiquitinase OTUB1 via a click reaction, to selectively stabilize tumor suppressor transcription factors. We developed three series of TF-DUBTACs, namely, FOXO-DUBTAC, p53-DUBTAC, and IRF-DUBTAC, which stabilize FOXO3A, p53, and IRF3 in cells, respectively, in an OTUB1-dependent manner. These results suggest that TF-DUBTAC is a generalizable platform to achieve selective stabilization of tumor suppressor transcription factors as a therapeutic means to suppress tumorigenesis.
Conflict of interest statement
The authors declare the following competing financial interest(s): W.W. is a co-founder and stockholder of the Rekindle Therapeutics. J.J. is a co-founder and equity shareholder in Cullgen, Inc. and a consultant for Cullgen, Inc., EpiCypher, Inc., and Accent Therapeutics, Inc. The Jin laboratory received re-search funds from Celgene Corporation, Levo Therapeutics, Cullgen, Inc. and Cullinan Oncology. All other authors declare no competing interests.
Figures
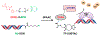
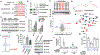
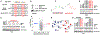

References
-
- Vogelstein B; Kinzler KW Cancer genes and the pathways they control. Nat. Med 2004, 10, 789–799. - PubMed
-
- Hanahan D; Weinberg RA Hallmarks of cancer: the next generation. Cell 2011, 144, 646–674. - PubMed
-
- Lehmann JM; Moore LB; Smith-Oliver TA; Wilkison WO; Willson TM; Kliewer SA An Antidiabetic Thiazolidinedione Is a High Affinity Ligand for Peroxisome Proliferator-activated Receptor γ (PPARγ). J. Biol. Chem 1995, 270, 12953–12956. - PubMed
-
- Willson TM; Cobb JE; Cowan DJ; Wiethe RW; Correa ID; Prakash SR; Beck KD; Moore LB; Kliewer SA; Lehmann JM The Structure–Activity Relationship between Peroxisome Proliferator-Activated Receptor γ Agonism and the Antihyperglycemic Activity of Thiazolidinediones. J. Med. Chem 1996, 39, 665–668. - PubMed
-
- Momose Y; Maekawa T; Yamano T; Kawada M; Odaka H; Ikeda H; Sohda T Novel 5-substituted 2,4-thiazolidinedione and 2,4-oxazolidinedione derivatives as insulin sensitizers with antidiabetic activities. J. Med. Chem 2002, 45, 1518–1534. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

