CRISPR screening uncovers a central requirement for HHEX in pancreatic lineage commitment and plasticity restriction
- PMID: 35787684
- PMCID: PMC9283336
- DOI: 10.1038/s41556-022-00946-4
CRISPR screening uncovers a central requirement for HHEX in pancreatic lineage commitment and plasticity restriction
Abstract
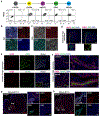
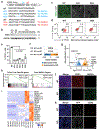
The pancreas and liver arise from a common pool of progenitors. However, the underlying mechanisms that drive their lineage diversification from the foregut endoderm are not fully understood. To tackle this question, we undertook a multifactorial approach that integrated human pluripotent-stem-cell-guided differentiation, genome-scale CRISPR-Cas9 screening, single-cell analysis, genomics and proteomics. We discovered that HHEX, a transcription factor (TF) widely recognized as a key regulator of liver development, acts as a gatekeeper of pancreatic lineage specification. HHEX deletion impaired pancreatic commitment and unleashed an unexpected degree of cellular plasticity towards the liver and duodenum fates. Mechanistically, HHEX cooperates with the pioneer TFs FOXA1, FOXA2 and GATA4, shared by both pancreas and liver differentiation programmes, to promote pancreas commitment, and this cooperation restrains the shared TFs from activating alternative lineages. These findings provide a generalizable model for how gatekeeper TFs like HHEX orchestrate lineage commitment and plasticity restriction in broad developmental contexts.
© 2022. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Competing interests
The authors declare no competing interests.
Figures











References
-
- Willnow D et al. Quantitative lineage analysis identifies a hepato-pancreato-biliary progenitor niche. Nature 597, 87–91 (2021). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

