The NCF1 variant p.R90H aggravates autoimmunity by facilitating the activation of plasmacytoid dendritic cells
- PMID: 35788118
- PMCID: PMC9374378
- DOI: 10.1172/JCI153619
The NCF1 variant p.R90H aggravates autoimmunity by facilitating the activation of plasmacytoid dendritic cells
Abstract
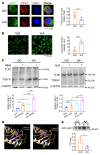
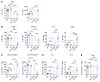
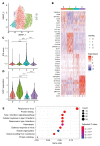
Plasmacytoid dendritic cells (pDCs) are a professional type I IFN producer that play critical roles in the pathogenesis of autoimmune diseases. However, both genetic regulation of the function of pDCs and their relationships with autoimmunity are largely undetermined. Here, we investigated the causality of the neutrophil cytosolic factor 1 (NCF1) missense variant, which is one of the most significant associated risk variants for lupus, and found that the substitution of arginine (R) for histidine (H) at position 90 in the NCF1 protein (NCF1 p.R90H) led to excessive activation of pDCs. A mechanism study demonstrated that p.R90H reduced the affinity of NCF1 for phospholipids, thereby impairing endosomal localization of NCF1. As NCF1 is a subunit of the NADPH oxidase 2 (NOX2) complex, this impairment led to an acidified endosomal pH and facilitated downstream TLR signaling. Consistently, the homozygous knockin mice manifested aggravated lupus progression in a pDC-dependent lupus model. More important, pharmaceutical intervention revealed that hydroxychloroquine (HCQ) could antagonize the detrimental function of NCF1 p.R90H in the lupus model and systemic lupus erythematosus samples, supporting the idea that NCF1 p.R90H could be identified as a genetic biomarker for HCQ application. Therefore, our study provides insights into the genetic control of pDC function and a paradigm for applying genetic variants to improve targeted therapy for autoimmune diseases.
Keywords: Autoimmunity; Lupus.
Figures





Comment in
-
Risk variant regulates pDC hyperactivation in SLE.Nat Rev Rheumatol. 2022 Sep;18(9):495. doi: 10.1038/s41584-022-00825-0. Nat Rev Rheumatol. 2022. PMID: 35927579 No abstract available.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

